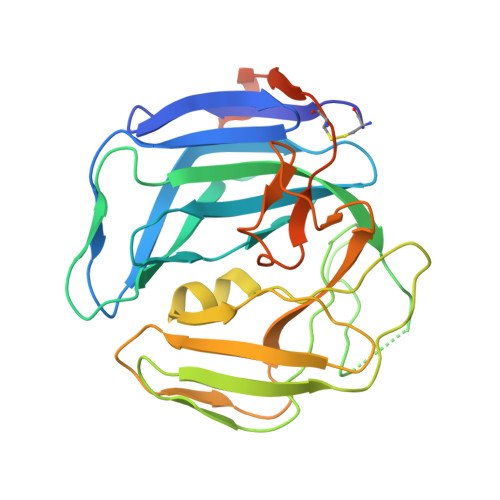
The Monomeric Dutpase from Epstein-Barr Virus Mimics Trimeric Dutpases
Tarbouriech, N., Buisson, M., Seigneurin, J.-M., Cusack, S., Burmeister, W.P.(2005) Structure 13: 1299
- PubMed: 16154087
- DOI: https://doi.org/10.1016/j.str.2005.06.009
- Primary Citation of Related Structures:
2BSY, 2BT1 - PubMed Abstract:
Deoxyuridine 5'-triphosphate pyrophosphatases (dUTPases) are ubiquitous enzymes cleaving dUTP into dUMP and pyrophosphate. They occur as monomeric, dimeric, or trimeric molecules. The trimeric and monomeric enzymes both contain the same five characteristic sequence motifs but in a different order, whereas the dimeric enzymes are not homologous. Monomeric dUTPases only occur in herpesviruses, such as Epstein-Barr virus (EBV). Here, we describe the crystal structures of EBV dUTPase in complex with the product dUMP and a substrate analog alpha,beta-imino-dUTP. The molecule consists of three domains forming one active site that has a structure extremely similar to one of the three active sites of trimeric dUTPases. The three domains functionally correspond to the subunits of the trimeric form. Domains I and II have the dUTPase fold, but they differ considerably in the regions that are not involved in the formation of the unique active site, whereas domain III has only little secondary structure.
- EMBL-Grenoble Outstation, BP181, F-38042 Grenoble cedex 9, France. tarbour@embl-grenoble.fr
Organizational Affiliation: