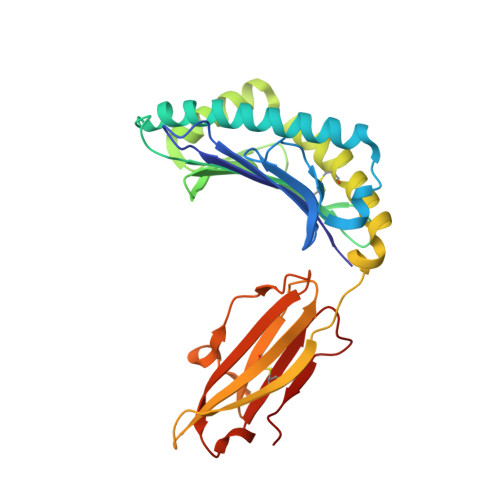
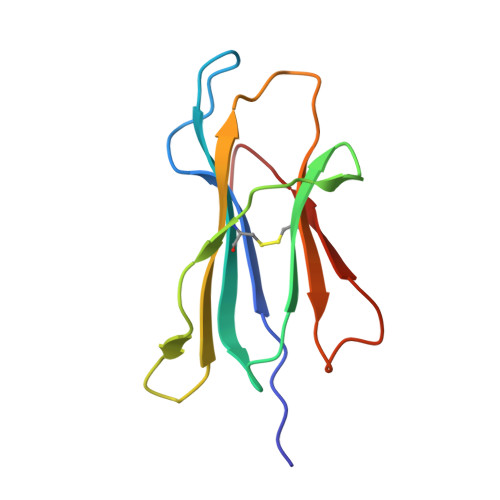
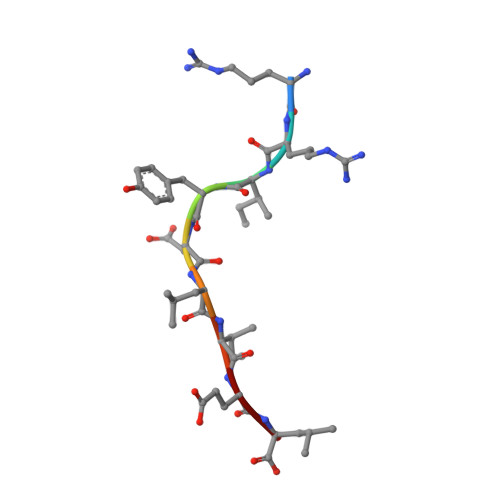
Crystal Structures and Kir3Dl1 Recognition of Three Immunodominant Viral Peptides Complexed to Hla-B2705
Stewart-Jones, G.B.E., Di Gleria, K., Kollnberger, S., Mcmichael, A.J., Jones, E.Y., Bowness, P.(2005) Eur J Immunol 35: 341
- PubMed: 15657948
- DOI: https://doi.org/10.1002/eji.200425724
- Primary Citation of Related Structures:
2BSR, 2BSS, 2BST - PubMed Abstract:
We have solved the crystal structures of three HLA-B*2705-peptide complexes with the immunodominant viral peptides: EBV EBNA3C 258-266 (RRIYDLIEL), influenza (flu) nucleoprotein NP383-391 (SRYWAIRTR), and HIV gag 264-273 (KRWIILGLNK). Long-term non-progression during HIV infection has been associated with presentation by HLA-B*2705, and T cell recognition, of the highly immunodominant KRWIILGLNK peptide. The tight hydrogen-bonding network observed between the HLA-B*2705 B-pocket and the peptide P2 arginine guanadinium anchor explains why mutation of this residue during HIV infection results in loss of peptide binding, immune escape and progression to AIDS. Prominent, solvent-exposed structures within these peptides may participate in generating T cell responses to these immunodominant epitopes. In the HLA-B*2705 complex with flu NP383-391, the amino acid side chains of residues 4, 7 and 8 are solvent-exposed whilst in the HIV decamer, the main-chain bulges into the solvent around P7. Thus, HLA-B*2705 presents viral peptides in a range of conformations. Tetrameric complexes of HLA-B*2705 with the HIV and flu but not EBV peptides bound strongly to the killer-Ig-like receptor (KIR)3DL1. Substitution of EBV P8 glutamate to threonine allowed recognition by KIR3DL1. In the HLA-B*2705-EBV structure the P8 glutamate side chain is solvent-exposed and may inhibit KIR3DL1 binding through electrostatic forces.
- The Division of Structural Biology, The Wellcome Trust Centre for Human Genetics, Oxford, UK.
Organizational Affiliation: