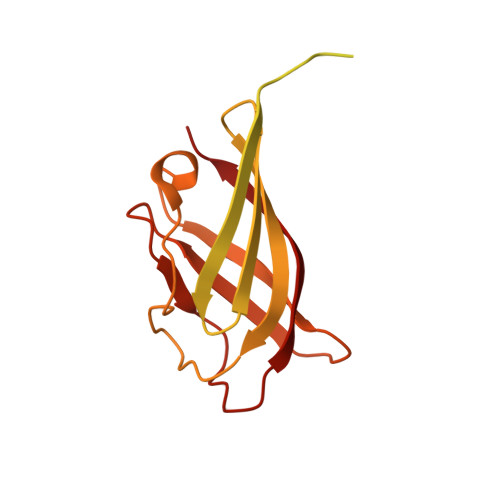
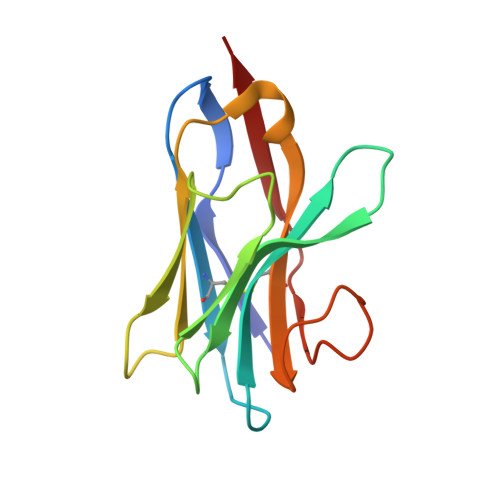
Lactococcal Bacteriophage P2 Receptor Binding Protein Structure Suggests a Common Ancestor Gene with Bacterial and Mammalian Viruses.
Spinelli, S., Desmyter, A., Verrips, C.T., de Haard, H.J.W., Moineau, S., Cambillau, C.(2006) Nat Struct Mol Biol 13: 85
- PubMed: 16327804
- DOI: https://doi.org/10.1038/nsmb1029
- Primary Citation of Related Structures:
2BSD, 2BSE - PubMed Abstract:
Lactococcus lactis is a Gram-positive bacterium used extensively by the dairy industry for the manufacture of fermented milk products. The double-stranded DNA bacteriophage p2 infects specific L. lactis strains using a receptor-binding protein (RBP) located at the tip of its noncontractile tail. We have solved the crystal structure of phage p2 RBP, a homotrimeric protein composed of three domains: the shoulders, a beta-sandwich attached to the phage; the neck, an interlaced beta-prism; and the receptor-recognition head, a seven-stranded beta-barrel. We used the complex of RBP with a neutralizing llama VHH domain to identify the receptor-binding site. Structural similarity between the recognition-head domain of phage p2 and those of adenoviruses and reoviruses, which invade mammalian cells, suggests that these viruses, despite evolutionary distant targets, lack of sequence similarity and the different chemical nature of their genomes (DNA versus RNA), might have a common ancestral gene.
- Architecture et Fonction des Macromolécules Biologiques, UMR 6098 CNRS and Universités d'Aix-Marseille I & II, Campus de Luminy, 163 Av. de Luminy 13288 Marseille Cedex 9, France.
Organizational Affiliation: