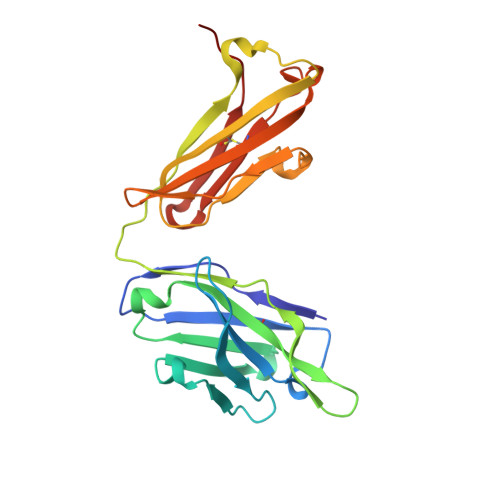
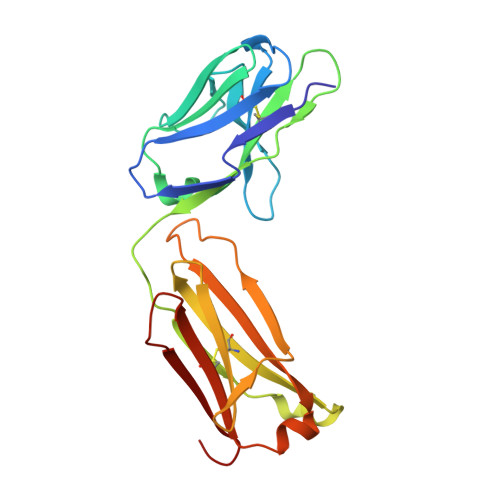
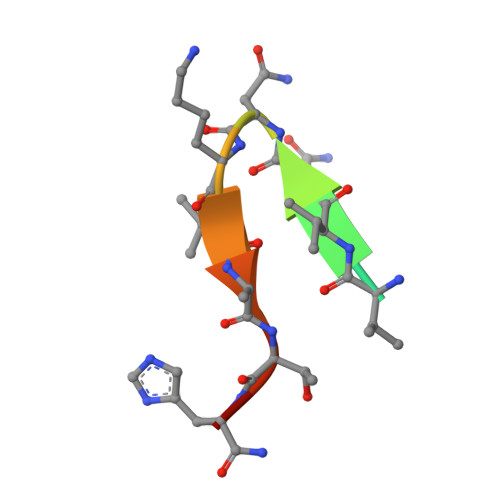
Crystal Structure of an Anti-Meningococcal Subtype P1.4 Pora Antibody Provides Basis for Peptide- Vaccine Design.
Oomen, C.J., Hoogerhout, P., Kuipers, B., Vidarsson, G., Van Alphen, L., Gros, P.(2005) J Mol Biology 351: 1070
- PubMed: 16038932
- DOI: https://doi.org/10.1016/j.jmb.2005.06.061
- Primary Citation of Related Structures:
2BRR - PubMed Abstract:
In various western countries, subtype P1.4 of Neisseria meningitidis serogroup B causes the greatest incidence of meningococcal disease. To investigate the molecular recognition of this subtype, we crystallised a peptide (P1HVVVNNKVATH(P11)), corresponding to the subtype P1.4 epitope sequence of outer membrane protein PorA, in complex with a Fab fragment of the bactericidal antibody MN20B9.34 directed against this epitope. Structure determination at 1.95 A resolution revealed a unique complex of one P1.4 antigen peptide bound to two identical Fab fragments. One Fab recognises the putative epitope residues in a 2:2 type I beta-turn at residues P5NNKV(P8), whereas the other Fab binds the C-terminal residues of the peptide that we consider a crystallisation artefact. Interestingly, recognition of the P1.4 epitope peptide is mediated almost exclusively through the complementarity-determining regions of the heavy chain. We exploited the observed turn conformation for designing conformationally restricted cyclic peptides for use as a peptide vaccine. The conformational stability of the two peptide designs was assessed by molecular dynamics simulations. Unlike the linear peptide, both cyclic peptides, conjugated to tetanus toxoid as a carrier protein, elicited antibody responses in mice that recognised meningococci of subtype P1.7-2,4. Serum bactericidal assays showed that some, but not all, of the sera induced with the cyclic peptide conjugates could activate the complement system with titres that were very high compared to the titres induced by complete PorA protein in its native conformation administered in outer membrane vesicles.
- Department of Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands.
Organizational Affiliation: