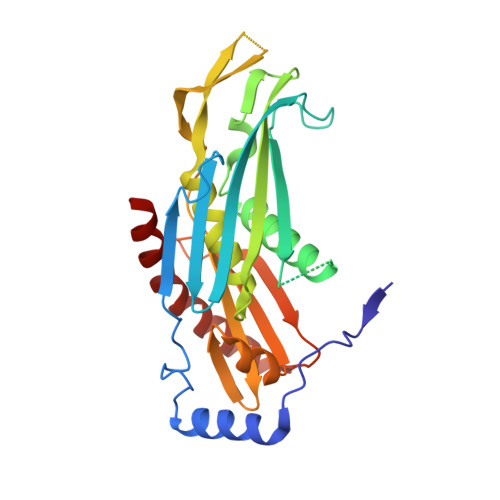
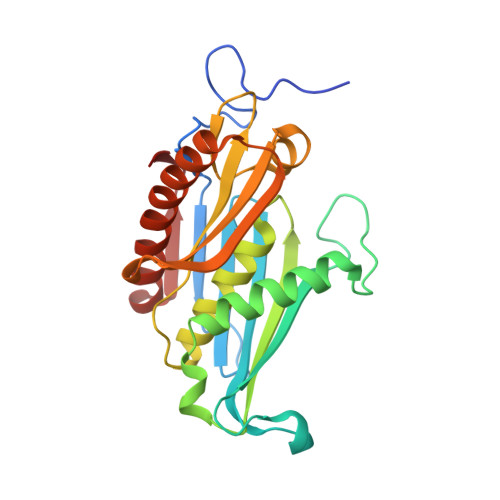
The Archaeal Exosome Core is a Hexameric Ring Structure with Three Catalytic Subunits.
Lorentzen, E., Walter, P., Fribourg, S., Evguenieva-Hackenberg, E., Klug, G., Conti, E.(2005) Nat Struct Mol Biol 12: 575
- PubMed: 15951817
- DOI: https://doi.org/10.1038/nsmb952
- Primary Citation of Related Structures:
2BR2 - PubMed Abstract:
The exosome is a 3' --> 5' exoribonuclease complex involved in RNA processing. We report the crystal structure of the RNase PH core complex of the Sulfolobus solfataricus exosome determined at a resolution of 2.8 A. The structure reveals a hexameric ring-like arrangement of three Rrp41-Rrp42 heterodimers, where both subunits adopt the RNase PH fold common to phosphorolytic exoribonucleases. Structure-guided mutagenesis reveals that the activity of the complex resides within the active sites of the Rrp41 subunits, all three of which face the same side of the hexameric structure. The Rrp42 subunit is inactive but contributes to the structuring of the Rrp41 active site. The high sequence similarity of this archaeal exosome to eukaryotic exosomes and its high structural similarity to the bacterial mRNA-degrading PNPase support a common basis for RNA-degrading machineries in all three domains of life.
- European Molecular Biology Laboratory, Heidelberg, Germany.
Organizational Affiliation: