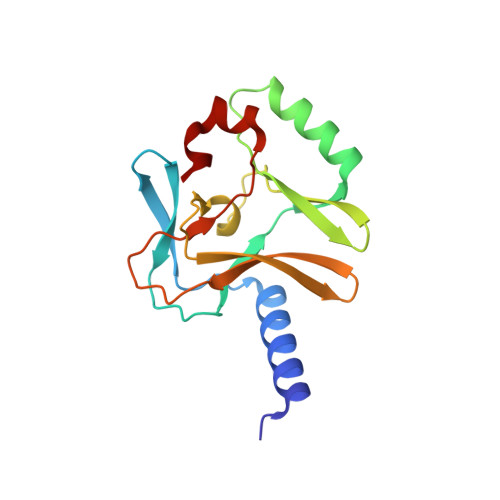
Specificity and Mechanism of the Histone Methyltransferase Pr-Set7
Xiao, B., Jing, C., Kelly, G., Walker, P.A., Muskett, F.W., Frenkiel, T.A., Martin, S.R., Sarma, K., Reinberg, D., Gamblin, S.J., Wilson, J.R.(2005) Genes Dev 19: 1444
- PubMed: 15933069
- DOI: https://doi.org/10.1101/gad.1315905
- Primary Citation of Related Structures:
2BQZ - PubMed Abstract:
Methylation of lysine residues of histones is an important epigenetic mark that correlates with functionally distinct regions of chromatin. We present here the crystal structure of a ternary complex of the enzyme Pr-Set7 (also known as Set8) that methylates Lys 20 of histone H4 (H4-K20). We show that the enzyme is exclusively a mono-methylase and is therefore responsible for a signaling role quite distinct from that established by other enzymes that target this histone residue. We provide evidence from NMR for the C-flanking domains of SET proteins becoming ordered upon addition of AdoMet cofactor and develop a model for the catalytic cycle of these enzymes. The crystal structure reveals the basis of the specificity of the enzyme for H4-K20 because a histidine residue within the substrate, close to the target lysine, is required for completion of the active site. We also show how a highly variable component of the SET domain is responsible for many of the enzymes' interactions with its target histone peptide and probably also how this part of the structure ensures that Pr-Set7 is nucleosome specific.
- National Institute for Medical Research, The Ridgeway, Mill Hill, London, NW7 1AA, United Kingdom.
Organizational Affiliation: