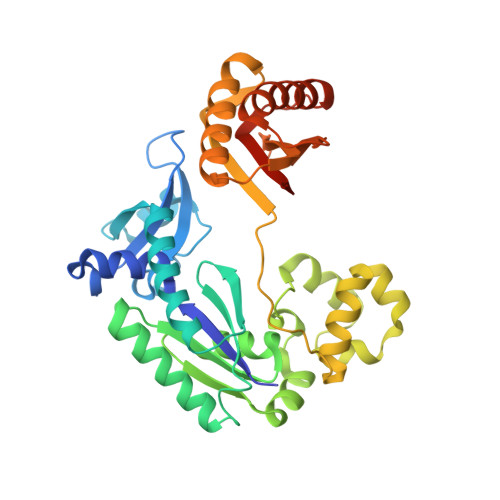
DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
Zang, H., Goodenough, A.K., Choi, J.Y., Irimia, A., Loukachevitch, L.V., Kozekov, I.D., Angel, K.C., Rizzo, C.J., Egli, M., Guengerich, F.P.(2005) J Biological Chem 280: 29750
- PubMed: 15965231
- DOI: https://doi.org/10.1074/jbc.M504756200
- Primary Citation of Related Structures:
2BQ3, 2BQR, 2BQU, 2BR0 - PubMed Abstract:
1,N(2)-Etheno(epsilon)guanine is a mutagenic DNA lesion derived from lipid oxidation products and also from some chemical carcinogens. Gel electrophoretic analysis of the products of primer extension by Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) indicated preferential incorporation of A opposite 3'-(1,N(2)-epsilon-G)TACT-5', among the four dNTPs tested individually. With the template 3'-(1,N(2)-epsilon-G)CACT-5', both G and A were incorporated. When primer extension was done in the presence of a mixture of all four dNTPs, high pressure liquid chromatography-mass spectrometry analysis of the products indicated that (opposite 3'-(1,N(2)-epsilon-G)CACT-5') the major product was 5'-GTGA-3' and the minor product was 5'-AGTGA-3'. With the template 3'-(1,N(2)-epsilon-G)TACT-5', the following four products were identified by high pressure liquid chromatography-mass spectrometry: 5'-AATGA-3', 5'-ATTGA-3', 5'-ATGA-3', and 5'-TGA-3'. An x-ray crystal structure of Dpo4 was solved (2.1 A) with a primer-template and A placed in the primer to be opposite the 1,N(2)-epsilon-G in the template 3'-(1,N(2)-epsilon-G)TACT 5'. The added A in the primer was paired across the template T with classic Watson-Crick geometry. Similar structures were observed in a ternary Dpo4-DNA-dATP complex and a ternary Dpo4-DNA-ddATP complex, with d(d)ATP opposite the template T. A similar structure was observed with a ddGTP adjacent to the primer and opposite the C next to 1,N(2)-epsilon-G in 3'-(1,N(2)-epsilon-G)CACT-5'. We concluded that Dpo4 uses several mechanisms, including A incorporation opposite 1,N(2)-epsilon-G and also a variation of dNTP-stabilized misalignment, to generate both base pair and frameshift mutations.
- Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, TN 37232, USA.
Organizational Affiliation: