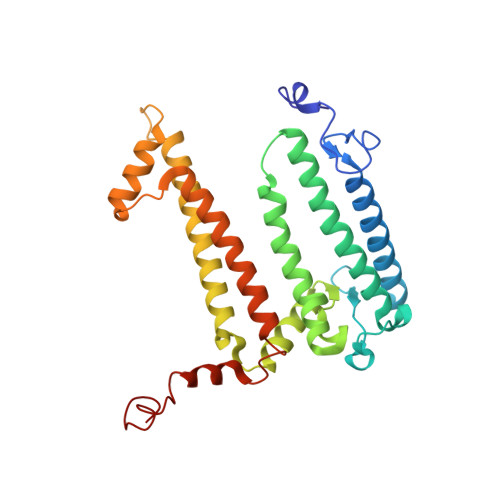
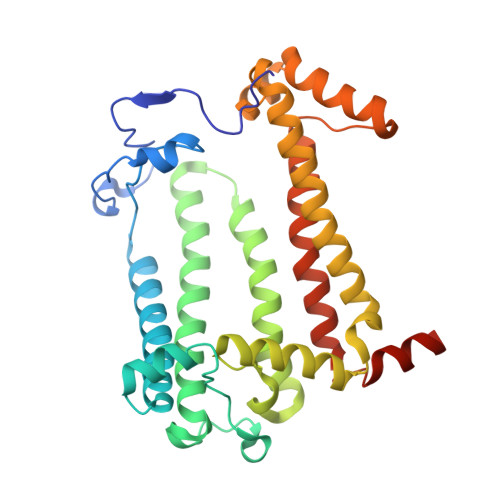
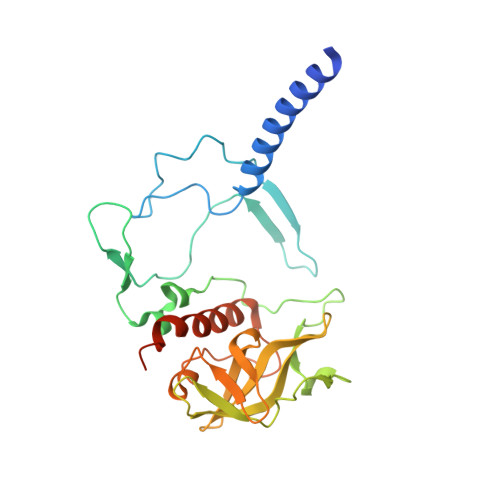
Conformational Regulation of Charge Recombination Reactions in a Photosynthetic Bacterial Reaction Centre
Katona, G., Snijder, A., Gourdon, P., Andreasson, U., Hansson, O., Andreasson, L.E., Neutze, R.(2005) Nat Struct Mol Biol 12: 630
- PubMed: 15937492
- DOI: https://doi.org/10.1038/nsmb948
- Primary Citation of Related Structures:
2BNP, 2BNS - PubMed Abstract:
In bright light the photosynthetic reaction center (RC) of Rhodobacter sphaeroides stabilizes the P(+)(870).Q(-)(A) charge-separated state and thereby minimizes the potentially harmful effects of light saturation. Using X-ray diffraction we report a conformational change that occurs within the cytoplasmic domain of this RC in response to prolonged illumination with bright light. Our observations suggest a novel structural mechanism for the regulation of electron transfer reactions in photosynthesis.
- Chalmers University of Technology, Department of Chemistry and Bioscience, PO Box 462, SE-405 30 Göteborg, Sweden.
Organizational Affiliation: