Structural and Kinetic Basis for Heightened Immunogenicity of T Cell Vaccines
Chen, J.-L., Stewart-Jones, G., Bossi, G., Lissin, N.M., Wooldridge, L., Choi, E.M.L., Held, G., Dunbar, P.R., Esnouf, R.M., Sami, M., Boultier, J.M., Rizkallah, P., Renner, C., Sewell, A., Van Der Merwe, P.A., Jackobsen, B.K., Griffiths, G., Jones, E.Y., Cerundolo, V.(2005) J Exp Medicine 201: 1243
- PubMed: 15837811
- DOI: https://doi.org/10.1084/jem.20042323
- Primary Citation of Related Structures:
2BNQ, 2BNR, 2BNU - PubMed Abstract:
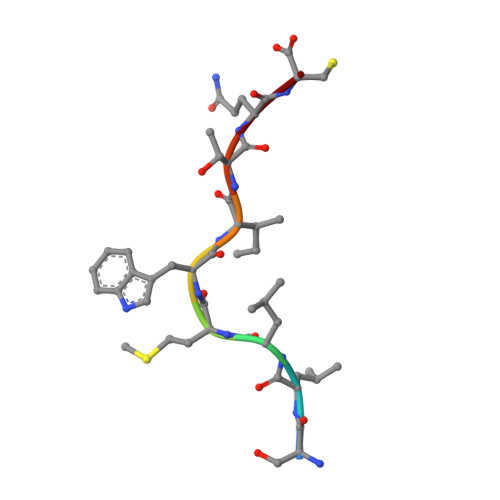
Analogue peptides with enhanced binding affinity to major histocompatibility class (MHC) I molecules are currently being used in cancer patients to elicit stronger T cell responses. However, it remains unclear as to how alterations of anchor residues may affect T cell receptor (TCR) recognition. We correlate functional, thermodynamic, and structural parameters of TCR-peptide-MHC binding and demonstrate the effect of anchor residue modifications of the human histocompatibility leukocyte antigens (HLA)-A2 tumor epitope NY-ESO-1(157-165)-SLLMWITQC on TCR recognition. The crystal structure of the wild-type peptide complexed with a specific TCR shows that TCR binding centers on two prominent, sequential, peptide sidechains, methionine-tryptophan. Cysteine-to-valine substitution at peptide position 9, while optimizing peptide binding to the MHC, repositions the peptide main chain and generates subtly enhanced interactions between the analogue peptide and the TCR. Binding analyses confirm tighter binding of the analogue peptide to HLA-A2 and improved soluble TCR binding. Recognition of analogue peptide stimulates faster polarization of lytic granules to the immunological synapse, reduces dependence on CD8 binding, and induces greater numbers of cross-reactive cytotoxic T lymphocyte to SLLMWITQC. These results provide important insights into heightened immunogenicity of analogue peptides and highlight the importance of incorporating structural data into the process of rational optimization of superagonist peptides for clinical trials.
- Tumour Immunology Unit, Weatherall Institute of Molecular Medicine, University of Oxford, John Radcliffe Hospital, Oxford OX3 9DS, UK.
Organizational Affiliation: