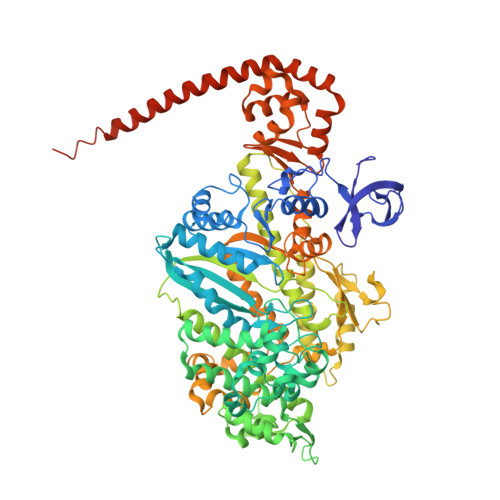
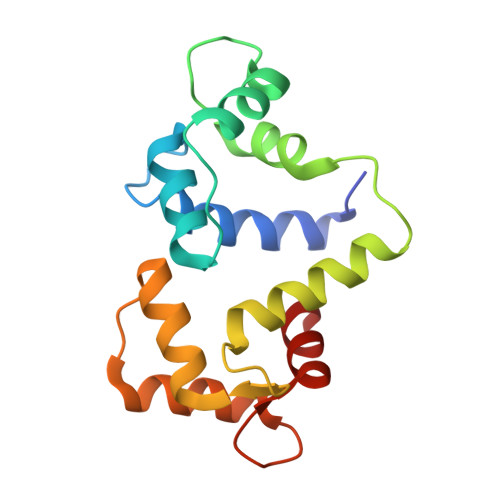
The Structure of the Myosin Vi Motor Reveals the Mechanism of Directionality Reversal
Menetrey, J., Bahloul, A., Wells, A., Yengo, C., Morris, C., Sweeney, H.L., Houdusse, A.(2005) Nature 435: 779
- PubMed: 15944696
- DOI: https://doi.org/10.1038/nature03592
- Primary Citation of Related Structures:
2BKH, 2BKI - PubMed Abstract:
Here we solve a 2.4-A structure of a truncated version of the reverse-direction myosin motor, myosin VI, that contains the motor domain and binding sites for two calmodulin molecules. The structure reveals only minor differences in the motor domain from that in plus-end directed myosins, with the exception of two unique inserts. The first is near the nucleotide-binding pocket and alters the rates of nucleotide association and dissociation. The second unique insert forms an integral part of the myosin VI converter domain along with a calmodulin bound to a novel target motif within the insert. This serves to redirect the effective 'lever arm' of myosin VI, which includes a second calmodulin bound to an 'IQ motif', towards the pointed (minus) end of the actin filament. This repositioning largely accounts for the reverse directionality of this class of myosin motors. We propose a model incorporating a kinesin-like uncoupling/docking mechanism to provide a full explanation of the movements of myosin VI.
- Structural Motility, Institut Curie CNRS, UMR144, 26 rue d'Ulm, 75248 Paris cedex 05, France.
Organizational Affiliation: