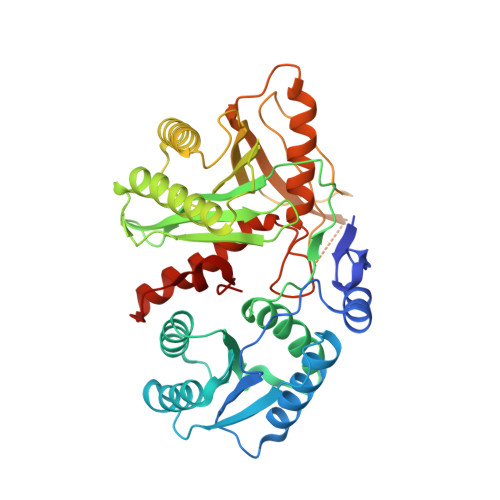
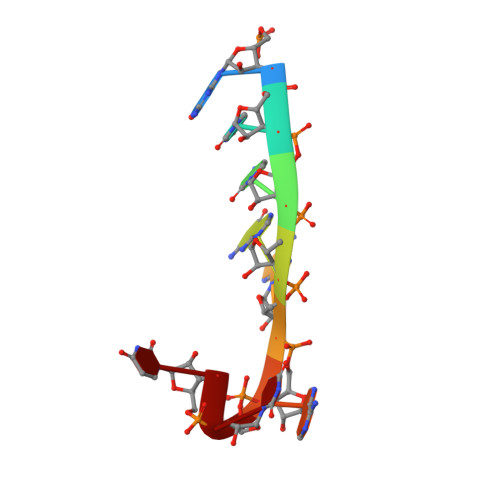
Structural Insights Into Mrna Recognition from a Piwi Domain-Sirna Guide Complex
Parker, J.S., Roe, S.M., Barford, D.(2005) Nature 434: 663
- PubMed: 15800628
- DOI: https://doi.org/10.1038/nature03462
- Primary Citation of Related Structures:
2BGG - PubMed Abstract:
RNA interference and related RNA silencing phenomena use short antisense guide RNA molecules to repress the expression of target genes. Argonaute proteins, containing amino-terminal PAZ (for PIWI/Argonaute/Zwille) domains and carboxy-terminal PIWI domains, are core components of these mechanisms. Here we show the crystal structure of a Piwi protein from Archaeoglobus fulgidus (AfPiwi) in complex with a small interfering RNA (siRNA)-like duplex, which mimics the 5' end of a guide RNA strand bound to an overhanging target messenger RNA. The structure contains a highly conserved metal-binding site that anchors the 5' nucleotide of the guide RNA. The first base pair of the duplex is unwound, separating the 5' nucleotide of the guide from the complementary nucleotide on the target strand, which exits with the 3' overhang through a short channel. The remaining base-paired nucleotides assume an A-form helix, accommodated within a channel in the PIWI domain, which can be extended to place the scissile phosphate of the target strand adjacent to the putative slicer catalytic site. This study provides insights into mechanisms of target mRNA recognition and cleavage by an Argonaute-siRNA guide complex.
- Section of Structural Biology, Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Road, London SW3 6JB, UK.
Organizational Affiliation: