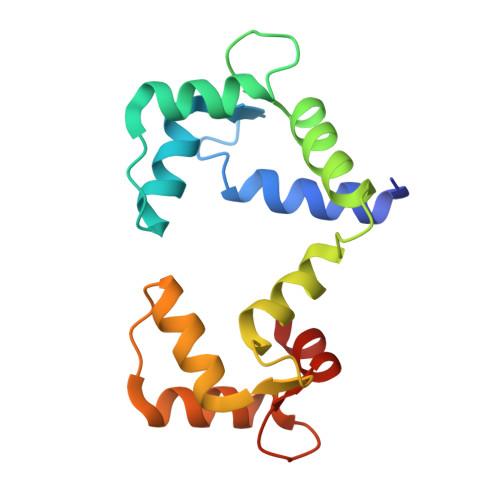
Complex of calmodulin with a ryanodine receptor target reveals a novel, flexible binding mode.
Maximciuc, A.A., Putkey, J.A., Shamoo, Y., Mackenzie, K.R.(2006) Structure 14: 1547-1556
- PubMed: 17027503
- DOI: https://doi.org/10.1016/j.str.2006.08.011
- Primary Citation of Related Structures:
2BCX - PubMed Abstract:
Calmodulin regulates ryanodine receptor-mediated Ca(2+) release through a conserved binding site. The crystal structure of Ca(2+)-calmodulin bound to this conserved site reveals that calmodulin recognizes two hydrophobic anchor residues at a novel "1-17" spacing that brings the calmodulin lobes close together but prevents them from contacting one another. NMR residual dipolar couplings demonstrate that the detailed structure of each lobe is preserved in solution but also show that the lobes experience domain motions within the complex. FRET measurements confirm the close approach of the lobes in binding the 1-17 target and show that calmodulin binds with one lobe to a peptide lacking the second anchor. We suggest that calmodulin regulates the Ca(2+) channel by switching between the contiguous binding mode seen in our crystal structure and a state where one lobe of calmodulin contacts the conserved binding site while the other interacts with a noncontiguous site on the channel.
- Department of Biochemistry and Cell Biology, Rice University, Houston, Texas 77005, USA.
Organizational Affiliation: