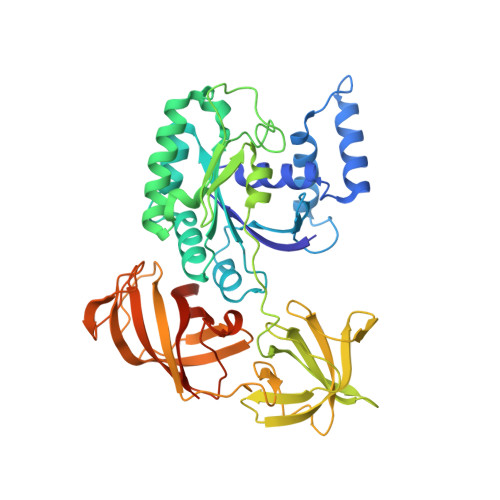
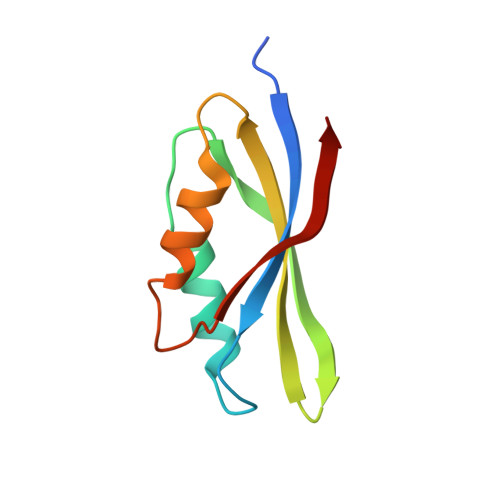
Mg2+ and a key lysine modulate exchange activity of eukaryotic translation elongation factor 1B alpha
Pittman, Y.R., Valente, L., Jeppesen, M.G., Andersen, G.R., Patel, S., Kinzy, T.G.(2006) J Biological Chem 281: 19457-19468
- PubMed: 16675455
- DOI: https://doi.org/10.1074/jbc.M601076200
- Primary Citation of Related Structures:
2B7B, 2B7C - PubMed Abstract:
To sustain efficient translation, eukaryotic elongation factor B alpha (eEF1B alpha) functions as the guanine nucleotide exchange factor for eEF1A. Stopped-flow kinetics using 2'-(or 3')-O-N-methylanthraniloyl (mant)-GDP showed spontaneous release of nucleotide from eEF1A is extremely slow and accelerated 700-fold by eEF1B alpha. The eEF1B alpha-stimulated reaction was inhibited by Mg2+ with a K(1/2) of 3.8 mM. Previous structural studies predicted the Lys-205 residue of eEF1B alpha plays an important role in promoting nucleotide exchange by disrupting the Mg2+ binding site. Co-crystal structures of the lethal K205A mutant in the catalytic C terminus of eEF1B alpha with eEF1A and eEF1A.GDP established that the lethality was not due to a structural defect. Instead, the K205A mutant drastically reduced the nucleotide exchange activity even at very low concentrations of Mg2+. A K205R eEF1B alpha mutant on the other hand was functional in vivo and showed nearly wild-type nucleotide dissociation rates but almost no sensitivity to Mg2+. These results indicate the significant role of Mg2+ in the nucleotide exchange reaction by eEF1B alpha and establish the catalytic function of Lys-205 in displacing Mg2+ from its binding site.
- Department of Molecular Genetics, Microbiology & Immunology, University of Medicine and Dentistry of New Jersey, Robert Wood Johnson Medical School, Piscataway 08854, USA.
Organizational Affiliation: