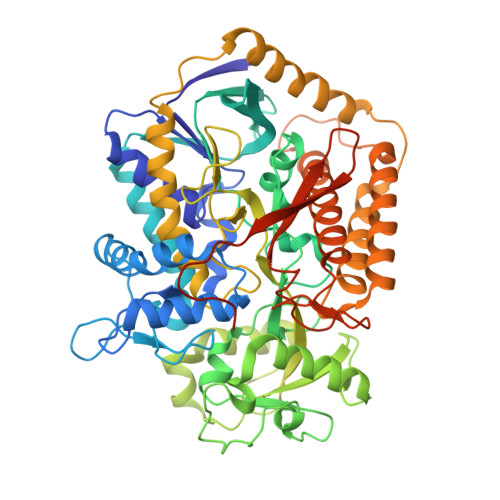
Fumarate Reductase and Succinate Oxidase Activity of Escherichia coli Complex II Homologs Are Perturbed Differently by Mutation of the Flavin Binding Domain
Maklashina, E., Iverson, T.M., Sher, Y., Kotlyar, V., Andrell, J., Mirza, O., Hudson, J.M., Armstrong, F.A., Rothery, R.A., Weiner, J.H., Cecchini, G.(2006) J Biological Chem 281: 11357-11365
- PubMed: 16484232
- DOI: https://doi.org/10.1074/jbc.M512544200
- Primary Citation of Related Structures:
2B76 - PubMed Abstract:
The Escherichia coli complex II homologues succinate:ubiquinone oxidoreductase (SQR, SdhCDAB) and menaquinol:fumarate oxidoreductase (QFR, FrdABCD) have remarkable structural homology at their dicarboxylate binding sites. Although both SQR and QFR can catalyze the interconversion of fumarate and succinate, QFR is a much better fumarate reductase, and SQR is a better succinate oxidase. An exception to the conservation of amino acids near the dicarboxylate binding sites of the two enzymes is that there is a Glu (FrdA Glu-49) near the covalently bound FAD cofactor in most QFRs, which is replaced with a Gln (SdhA Gln-50) in SQRs. The role of the amino acid side chain in enzymes with Glu/Gln/Ala substitutions at FrdA Glu-49 and SdhA Gln-50 has been investigated in this study. The data demonstrate that the mutant enzymes with Ala substitutions in either QFR or SQR remain functionally similar to their wild type counterparts. There were, however, dramatic changes in the catalytic properties when Glu and Gln were exchanged for each other in QFR and SQR. The data show that QFR and SQR enzymes are more efficient succinate oxidases when Gln is in the target position and a better fumarate reductase when Glu is present. Overall, structural and catalytic analyses of the FrdA E49Q and SdhA Q50E mutants suggest that coulombic effects and the electronic state of the FAD are critical in dictating the preferred directionality of the succinate/fumarate interconversions catalyzed by the complex II superfamily.
- Molecular Biology Division, Veterans Affairs Medical Center, San Francisco, California 94121, USA.
Organizational Affiliation: