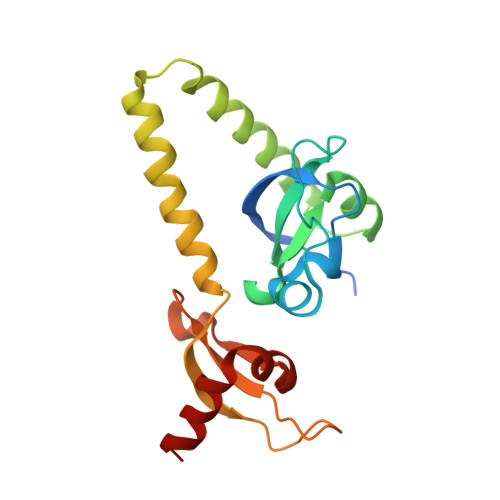
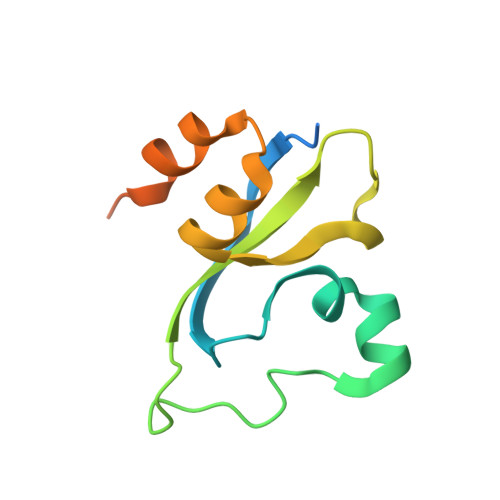
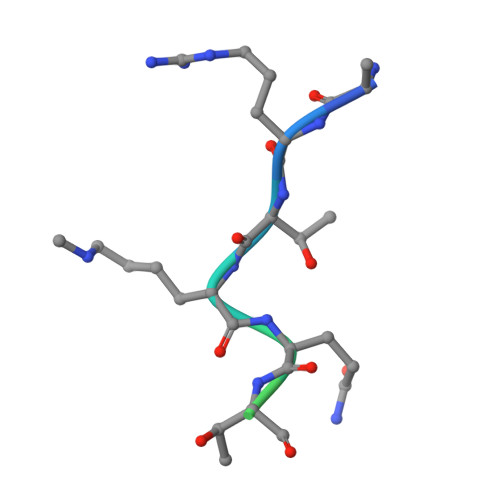
Double chromodomains cooperate to recognize the methylated histone H3 tail.
Flanagan, J.F., Mi, L.Z., Chruszcz, M., Cymborowski, M., Clines, K.L., Kim, Y., Minor, W., Rastinejad, F., Khorasanizadeh, S.(2005) Nature 438: 1181-1185
- PubMed: 16372014
- DOI: https://doi.org/10.1038/nature04290
- Primary Citation of Related Structures:
2B2T, 2B2U, 2B2V, 2B2W, 2B2Y - PubMed Abstract:
Chromodomains are modules implicated in the recognition of lysine-methylated histone tails and nucleic acids. CHD (for chromo-ATPase/helicase-DNA-binding) proteins regulate ATP-dependent nucleosome assembly and mobilization through their conserved double chromodomains and SWI2/SNF2 helicase/ATPase domain. The Drosophila CHD1 localizes to the interbands and puffs of the polytene chromosomes, which are classic sites of transcriptional activity. Other CHD isoforms (CHD3/4 or Mi-2) are important for nucleosome remodelling in histone deacetylase complexes. Deletion of chromodomains impairs nucleosome binding and remodelling by CHD proteins. Here we describe the structure of the tandem arrangement of the human CHD1 chromodomains, and its interactions with histone tails. Unlike HP1 and Polycomb proteins that use single chromodomains to bind to their respective methylated histone H3 tails, the two chromodomains of CHD1 cooperate to interact with one methylated H3 tail. We show that the human CHD1 double chromodomains target the lysine 4-methylated histone H3 tail (H3K4me), a hallmark of active chromatin. Methylammonium recognition involves two aromatic residues, not the three-residue aromatic cage used by chromodomains of HP1 and Polycomb proteins. Furthermore, unique inserts within chromodomain 1 of CHD1 block the expected site of H3 tail binding seen in HP1 and Polycomb, instead directing H3 binding to a groove at the inter-chromodomain junction.
- Department of Biochemistry and Molecular Genetics, University of Virginia Health System, Charlottesville, Virginia 22908, USA.
Organizational Affiliation: