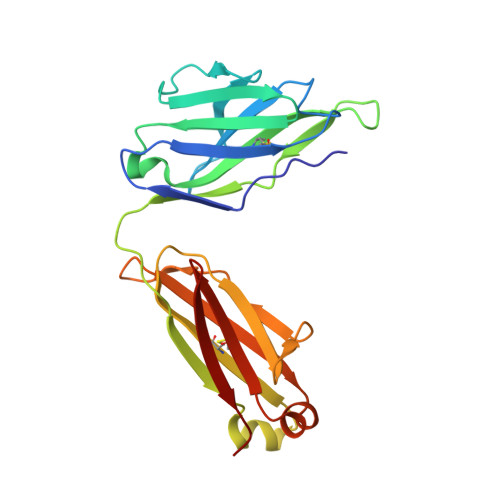
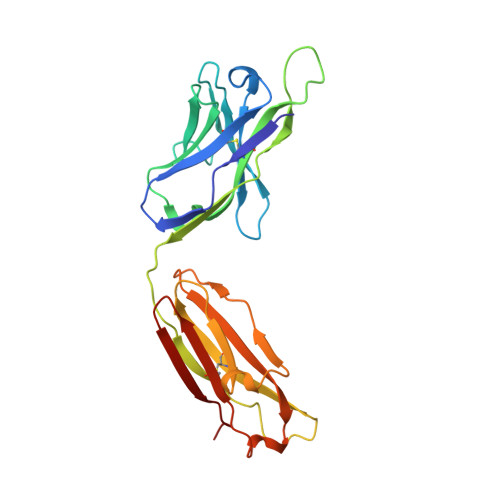
Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity.
Stanfield, R.L., Gorny, M.K., Zolla-Pazner, S., Wilson, I.A.(2006) J Virol 80: 6093-6105
- PubMed: 16731948
- DOI: https://doi.org/10.1128/JVI.00205-06
- Primary Citation of Related Structures:
2B0S, 2B1A, 2B1H - PubMed Abstract:
Human monoclonal antibody 2219 is a neutralizing antibody isolated from a human immunodeficiency virus type 1-infected individual. 2219 was originally selected for binding to a V3 fusion protein and can neutralize primary isolates from subtypes B, A, and F. Thus, 2219 represents a cross-reactive, human anti-V3 antibody. Fab 2219 binds to one face of the variable V3 beta-hairpin, primarily contacting conserved residues on the N-terminal beta-strand of V3, leaving the V3 crown or tip largely accessible. Three V3/2219 complexes reveal the antibody-bound conformations for both the N- and C-terminal regions that flank the V3 crown and illustrate how twisting of the V3 loop alters the relative dispositions and pairing of the amino acids in the adjacent V3 beta-strands and how the antibody can accommodate V3 loops with different sequences.
- Department of Molecular Biology, The Scripps Research Institute, La Jolla, CA 92037, USA. robyn@scripps.edu
Organizational Affiliation: