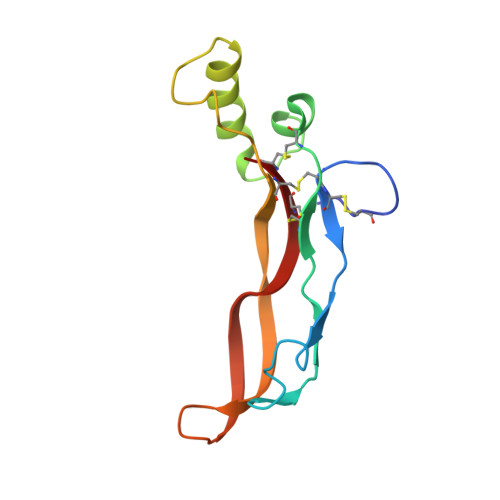
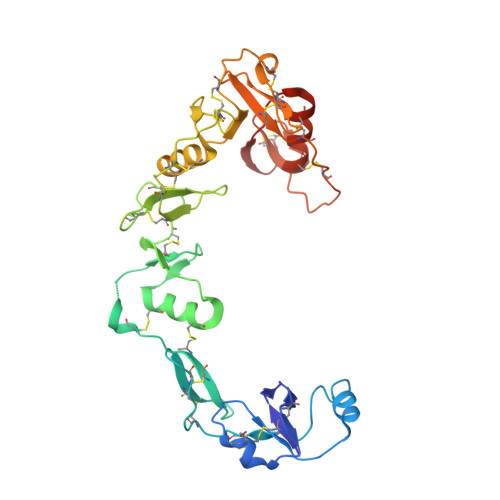
The Structure of the Follistatin:Activin Complex Reveals Antagonism of Both Type I and Type II Receptor Binding.
Thompson, T.B., Lerch, T.F., Cook, R.W., Woodruff, T.K., Jardetzky, T.S.(2005) Dev Cell 9: 535-543
- PubMed: 16198295
- DOI: https://doi.org/10.1016/j.devcel.2005.09.008
- Primary Citation of Related Structures:
2B0U - PubMed Abstract:
TGF-beta ligands stimulate diverse cellular differentiation and growth responses by signaling through type I and II receptors. Ligand antagonists, such as follistatin, block signaling and are essential regulators of physiological responses. Here we report the structure of activin A, a TGF-beta ligand, bound to the high-affinity antagonist follistatin. Two follistatin molecules encircle activin, neutralizing the ligand by burying one-third of its residues and its receptor binding sites. Previous studies have suggested that type I receptor binding would not be blocked by follistatin, but the crystal structure reveals that the follistatin N-terminal domain has an unexpected fold that mimics a universal type I receptor motif and occupies this receptor binding site. The formation of follistatin:BMP:type I receptor complexes can be explained by the stoichiometric and geometric arrangement of the activin:follistatin complex. The mode of ligand binding by follistatin has important implications for its ability to neutralize homo- and heterodimeric ligands of this growth factor family.
- Department of Biochemistry, Northwestern University, Evanston, Illinois 60208, USA.
Organizational Affiliation: