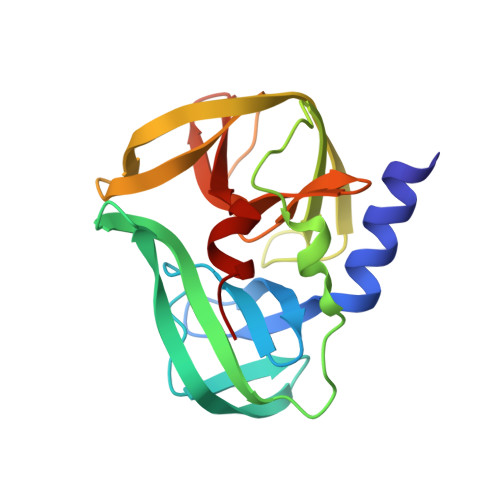
NMR solution structures of the apo and peptide-inhibited human rhinovirus 3C protease (Serotype 14): structural and dynamic comparison
Bjorndahl, T.C., Andrew, L.C., Semenchenko, V., Wishart, D.S.(2007) Biochemistry 46: 12945-12958
- PubMed: 17944485
- DOI: https://doi.org/10.1021/bi7010866
- Primary Citation of Related Structures:
2B0F, 2IN2 - PubMed Abstract:
The human rhinovirus (HRV) is a positive sense RNA virus responsible for about 30% of "common colds". It relies on a 182 residue cysteine protease (3C) to proteolytically process its single gene product. Inhibition of this enzyme in vitro and in vivo has consistently demonstrated cessation of viral replication. This suggests that 3C protease inhibitors could serve as good drug candidates. However, significant proteolytic substrate diversity exists within the 110+ known rhinovirus serotypes. To investigate this variability we used NMR to solve the structure of the rhinovirus serotype 14 3C protease (subgenus B) covalently bound to a peptide (acetyl-LEALFQ-ethylpropionate) inhibitor. The inhibitor-bound structure was determined to an overall rmsd of 0.82 A (backbone atoms) and 1.49 A (all heavy atoms). Comparison with the X-ray structure of the serotype 2 HRV 3C protease from subgenus A (51% sequence identity) bound to the inhibitor ruprintrivir allowed the identification of conserved intermolecular interactions involved in proximal substrate binding as well as subgenus differences that might account for the variability observed in SAR studies. To better characterize the 3C protease and investigate the structural and dynamic differences between the apo and bound states we also solved the solution structure of the apo form. The apo structure has an overall rmsd of 1.07 +/- 0.17 A over backbone atoms, which is greater by 0.25 A than what is seen for the inhibited enzyme (2B0F.pdb). This increase is localized to the enzyme's C-terminal beta-barrel domain, which is responsible for recognizing and binding proteolytic substrates. Amide hydrogen exchange dynamics revealed dramatic differences between the two enzyme states. Furthermore, a number of residues exhibited exchange-broadened amide NMR signals in the apo state compared to the inhibited state. The majority of these residues are associated with proteolytic substrate interaction.
- Faculty of Pharmacy and Pharmaceutical Sciences, University of Alberta, Edmonton, Alberta, Canada.
Organizational Affiliation: