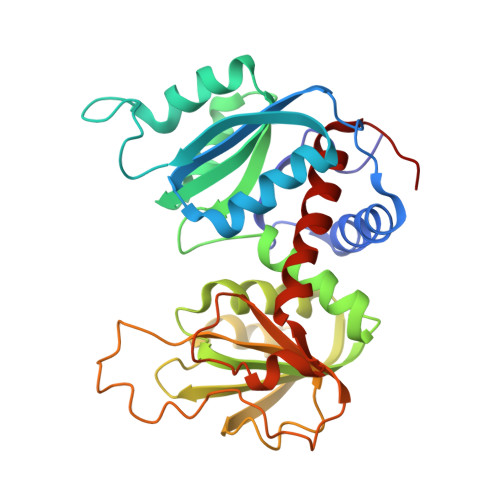
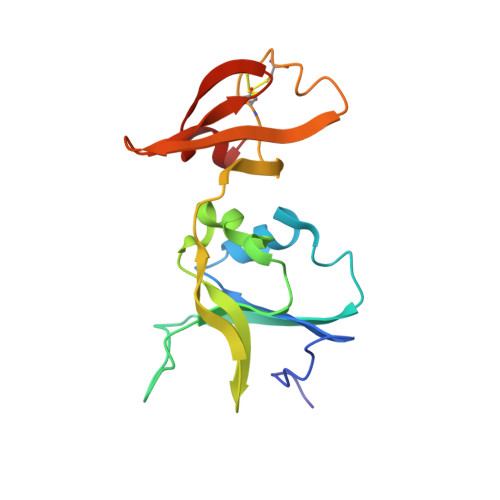
Crystal and molecular structures of native and CTP-liganded aspartate carbamoyltransferase from Escherichia coli.
Honzatko, R.B., Crawford, J.L., Monaco, H.L., Ladner, J.E., Ewards, B.F., Evans, D.R., Warren, S.G., Wiley, D.C., Ladner, R.C., Lipscomb, W.N.(1982) J Mol Biology 160: 219-263
- PubMed: 6757446
- DOI: https://doi.org/10.1016/0022-2836(82)90175-9
- Primary Citation of Related Structures:
2ATC