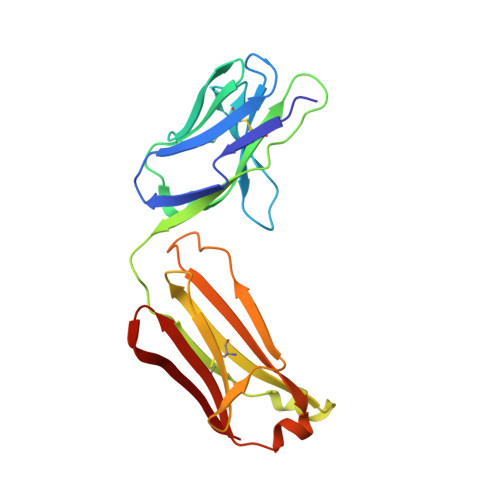
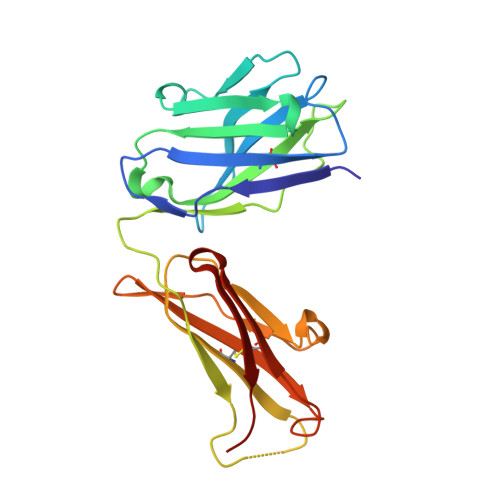
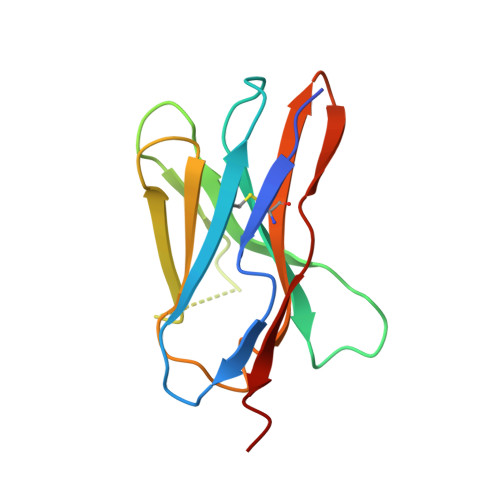
Crystal structure of the TCR co-receptor CD8alphaalpha in complex with monoclonal antibody YTS 105.18 Fab fragment at 2.88 A resolution.
Shore, D.A., Teyton, L., Dwek, R.A., Rudd, P.M., Wilson, I.A.(2006) J Mol Biology 358: 347-354
- PubMed: 16530222
- DOI: https://doi.org/10.1016/j.jmb.2006.02.016
- Primary Citation of Related Structures:
2ARJ - PubMed Abstract:
The CD8 glycoprotein functions as an essential element in the control of T-cell selection, maturation and the TCR-mediated response to peptide antigen. CD8 is expressed as both heterodimeric CD8alphabeta and homodimeric CD8alphaalpha isoforms, which have distinct physiological roles and exhibit tissue-specific expression patterns. CD8alphaalpha has previously been crystallized in complex with class I pMHC and, more recently, with the mouse class Ib thymic leukemia antigen (TL). Here, we present the crystal structure of a soluble form of mouse CD8alphaalpha in complex with rat monoclonal antibody YTS 105.18 Fab fragment at 2.88 A resolution. YTS 105.18, which is commonly used in the blockade of CD8+ T-cell activation in response to peptide antigen, is specific for mouse CD8alpha. The YTS 105.18 Fab is one of only five rat IgG Fab structures to have been reported to date. Analysis of the YTS 105.18 Fab epitope on CD8alpha reveals that this antibody blocks CD8 activity by hydrogen bonding to residues that are critical for interaction with both class I pMHC and TL. Structural comparison of the liganded and unliganded forms of soluble CD8alphaalpha indicates that the mouse CD8alphaalpha immunoglobulin-domain dimer does not undergo significant structural alteration upon interaction either with class I pMHC or TL.
- Department of Molecular Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: