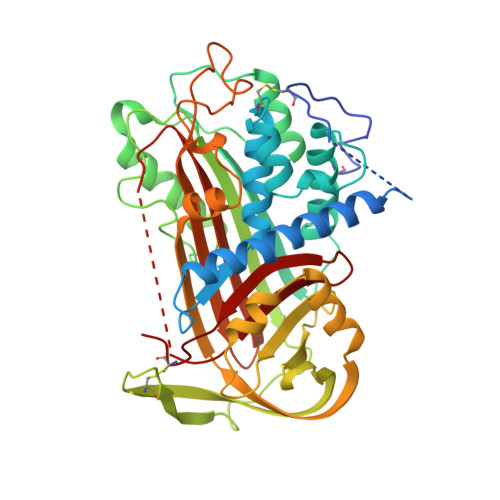
The 2.6 A structure of antithrombin indicates a conformational change at the heparin binding site.
Skinner, R., Abrahams, J.P., Whisstock, J.C., Lesk, A.M., Carrell, R.W., Wardell, M.R.(1997) J Mol Biology 266: 601-609
- PubMed: 9067613
- DOI: https://doi.org/10.1006/jmbi.1996.0798
- Primary Citation of Related Structures:
2ANT - PubMed Abstract:
The crystal structure of a dimeric form of intact antithrombin has been solved to 2.6 A, representing the highest-resolution structure of an active, inhibitory serpin to date. The crystals were grown under microgravity conditions on Space Shuttle mission STS-67. The overall confidence in the structure, determined earlier from lower resolution data, is increased and new insights into the structure-function relationship are gained. Clear and continuous electron density is present for the reactive centre loop region P12 to P14 inserting into the top of the A-beta-sheet. Areas of the extended amino terminus, unique to antithrombin and important in the binding of the glycosaminoglycan heparin, can now be traced further than in the earlier structures. As in the earlier studies, the crystals contain one active and one latent molecule per asymmetric unit. Better definition of the electron density surrounding the D-helix and of the residues implicated in the binding of the heparin pentasaccharide (Arg47, Lys114, Lys125, Arg129) provides an insight into the change of affinity of binding that accompanies the change in conformation. In particular, the observed hydrogen bonding of these residues to the body of the molecule in the latent form explains the mechanism for the release of newly formed antithrombin-protease complexes into the circulation for catabolic removal.
- Department of Haematology, University of Cambridge, MRC Centre, UK.
Organizational Affiliation: