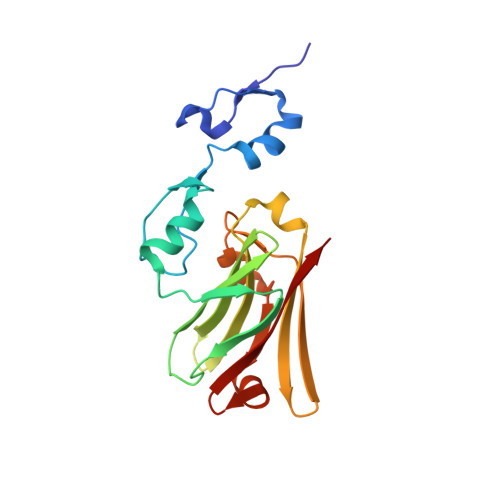
Elucidation of the substrate binding site of Siah ubiquitin ligase
House, C.M., Hancock, N.C., Moller, A., Cromer, B.A., Fedorov, V., Bowtell, D.D.L., Parker, M.W., Polekhina, G.(2006) Structure 14: 695-701
- PubMed: 16615911
- DOI: https://doi.org/10.1016/j.str.2005.12.013
- Primary Citation of Related Structures:
2AN6 - PubMed Abstract:
The Siah family of RING proteins function as ubiquitin ligase components, contributing to the degradation of multiple targets involved in cell growth, differentiation, angiogenesis, oncogenesis, and inflammation. Previously, a binding motif (degron) was recognized in many of the Siah degradation targets, suggesting that Siah itself may facilitate substrate recognition. We report the crystal structure of the Siah in complex with a peptide containing the degron motif. Binding is within a groove formed in part by the zinc fingers and the first two beta strands of the TRAF-C domain of Siah. We show that residues in the degron, previously described to facilitate binding to Siah, interact with the protein. Mutagenesis of Siah at sites of interaction also abrogates both in vitro peptide binding and destabilization of a known Siah target.
- Ian Potter Foundation Centre for Cancer Genomics and Predictive Medicine, Peter MacCallum Cancer Centre, Locked Bag 1 A'Beckett Street, Melbourne, Victoria 8006, Australia.
Organizational Affiliation: