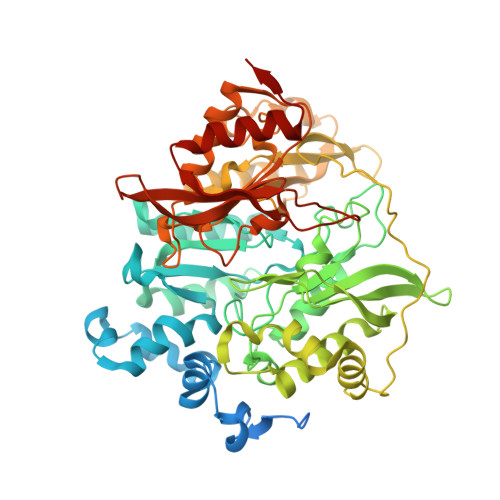
Structure of Spinach Nitrite Reductase: Implications for Multi-electron Reactions by the Iron-Sulfur:Siroheme Cofactor
Swamy, U., Wang, M., Tripathy, J.N., Kim, S.K., Hirasawa, M., Knaff, D.B., Allen, J.P.(2005) Biochemistry 44: 16054-16063
- PubMed: 16331965
- DOI: https://doi.org/10.1021/bi050981y
- Primary Citation of Related Structures:
2AKJ - PubMed Abstract:
The structure of nitrite reductase, a key enzyme in the process of nitrogen assimilation, has been determined using X-ray diffraction to a resolution limit of 2.8 A. The protein has a globular fold consisting of 3 alpha/beta domains with the siroheme-iron sulfur cofactor at the interface of the three domains. The Fe(4)S(4) cluster is coordinated by cysteines 441, 447, 482, and 486. The siroheme is located at a distance of 4.2 A from the cluster, and the central iron atom is coordinated to Cys 486. The siroheme is surrounded by several ionizable amino acid residues that facilitate the binding and subsequent reduction of nitrite. A model for the ferredoxin:nitrite reductase complex is proposed in which the binding of ferredoxin to a positively charged region of nitrite reductase results in elimination of exposure of the cofactors to the solvent. The structure of nitrite reductase shows a broad similarity to the hemoprotein subunit of sulfite reductase but has many significant differences in the backbone positions that could reflect sequence differences or could arise from alterations of the sulfite reductase structure that arise from the isolation of this subunit from the native complex. The implications of the nitrite reductase structure for understanding multi-electron processes are discussed in terms of differences in the protein environments of the cofactors.
- Department of Chemistry and Biochemistry, Arizona State University, Tempe Arizona 85287-1604, USA.
Organizational Affiliation: