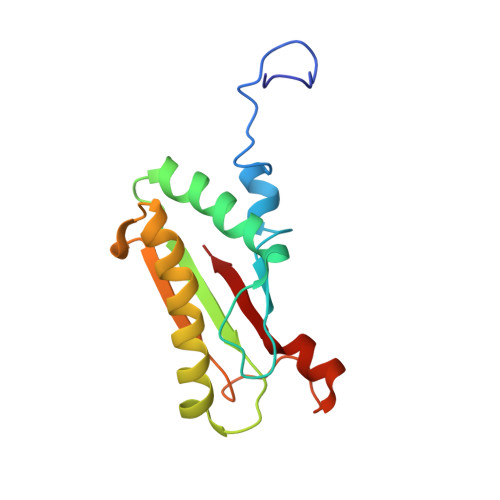
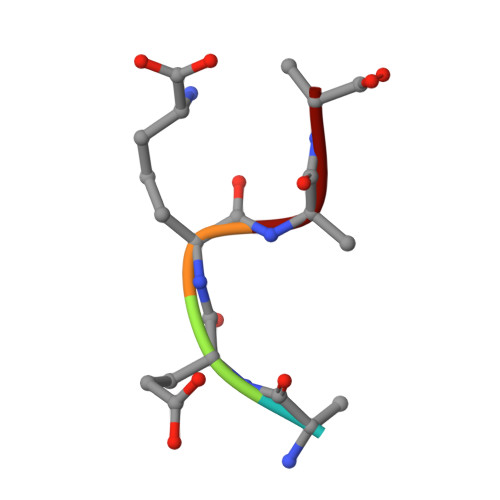
Peptidoglycan recognition by pal, an outer membrane lipoprotein.
Parsons, L.M., Lin, F., Orban, J.(2006) Biochemistry 45: 2122-2128
- PubMed: 16475801
- DOI: https://doi.org/10.1021/bi052227i
- Primary Citation of Related Structures:
2AIZ - PubMed Abstract:
Peptidoglycan-associated lipoprotein (Pal) is a potential vaccine candidate from Haemophilus influenzae that is highly conserved in Gram-negative bacteria and anchored to the outer membrane through an N-terminal lipid attachment. Pal stabilizes the outer membrane by providing a noncovalent link to the peptidoglycan (PG) layer through a periplasmic domain. Using NMR spectroscopy, we determined the three-dimensional structure of a complex between the periplasmic domain of Pal and a biosynthetic peptidoglycan precursor (PG-P), UDP-N-acetylmuramyl-L-Ala-alpha-d-Glu-m-Dap-D-Ala-d-Ala (m-Dap is meso-diaminopimelate). Pal has a binding pocket lined with conserved surface residues that interacts exclusively with the peptide portion of the ligand. The m-Dap residue, which is mainly found in the cell walls of Gram-negative bacteria, is sequestered in this pocket and plays an important role by forming hydrogen bond and hydrophobic contacts to Pal. The structure provides insight into the mode of cell wall recognition for a broad class of Gram-negative membrane proteins, including OmpA and MotB, which have peptidoglycan-binding domains homologous to that of Pal.
- Center for Advanced Research in Biotechnology, University of Maryland Biotechnology Institute, 9600 Gudelsky Drive, Rockville, Maryland 20850, USA.
Organizational Affiliation: