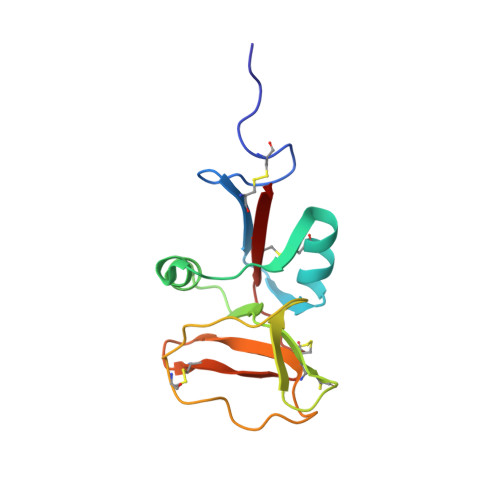
The solution structure of type II antifreeze protein reveals a new member of the lectin family.
Gronwald, W., Loewen, M.C., Lix, B., Daugulis, A.J., Sonnichsen, F.D., Davies, P.L., Sykes, B.D.(1998) Biochemistry 37: 4712-4721
- PubMed: 9537986
- DOI: https://doi.org/10.1021/bi972788c
- Primary Citation of Related Structures:
2AFP - PubMed Abstract:
A recombinant form of the sea raven type II antifreeze protein (SRAFP) has been produced using the Pichia pastoris expression system. The antifreeze activity of recombinant SRAFP is indistinguishable from that of the wild-type protein. The global fold of SRAFP has been determined by two-dimensional 1H homonuclear and three-dimensional 1H-¿15N¿ heteronuclear NMR spectroscopy using 785 NOE distance restraints and 47 angular restraints. The molecule folds into one globular domain that consists of two helices and nine beta-strands in two beta-sheets. The structure confirms the proposed existence of five disulfide bonds. The global fold of SRAFP is homologous to C-type lectins and pancreatic stone proteins, even though the sequence identity is only approximately 20%.
- Protein Engineering Network of Centres of Excellence, University of Alberta, Edmonton, Alberta T6G 2S2, Canada.
Organizational Affiliation: