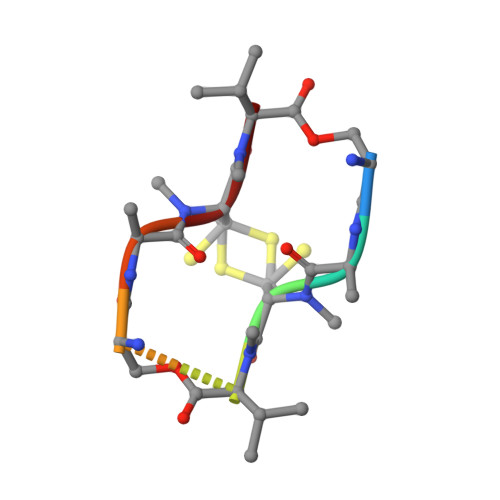
Serendipitous Sad Phasing of an Echinomycin-(Acgtacgt)2 Bisintercalation Complex.
Cuesta-Seijo, J.A., Weiss, M.S., Sheldrick, G.M.(2006) Acta Crystallogr D Biol Crystallogr 62: 417
- PubMed: 16552143
- DOI: https://doi.org/10.1107/S0907444906003763
- Primary Citation of Related Structures:
2ADW - PubMed Abstract:
A new crystal form was obtained for the complex between (ACGTACGT)2 and echinomycin and X-ray data were collected to 1.6 A. The structure was phased by the SAD method based on a single unexpected anomalous scatterer that could be identified as a mixture of nickel and zinc by measurements of the anomalous scattering at different wavelengths. This cation is coordinated by two guanines from two different duplexes and four water molecules. The structure resembles previously reported crystal structures of DNA-echinomycin complexes, except that three of the eight base pairs flanking the echinomycin bisintercalator sites have the Watson-Crick rather than the Hoogsteen configuration. Hoogsteen binding was found for the corresponding base pairs of the crystallographically independent duplex, indicating that the two configurations are very close in energy.
- Department of Structural Chemistry, University of Göttingen, Tammannstrasse 4, D37077 Göttingen, Germany.
Organizational Affiliation: