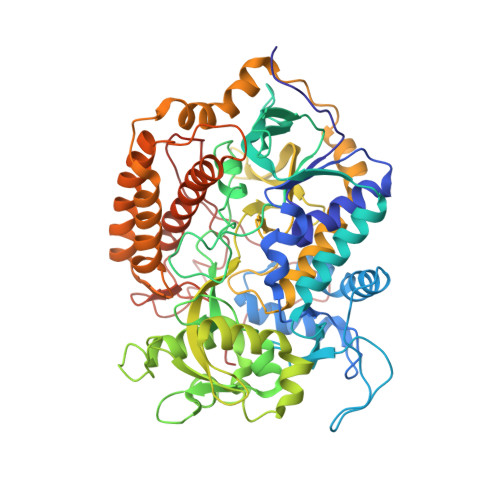
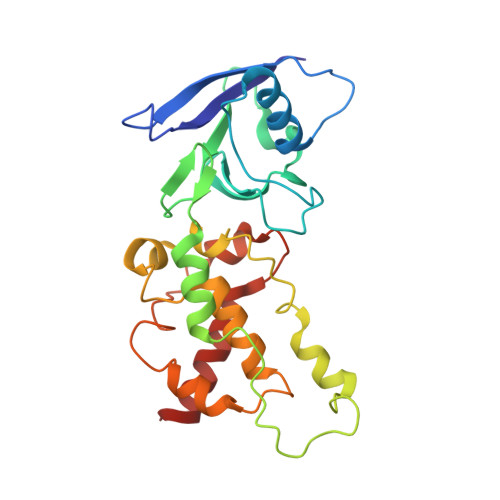
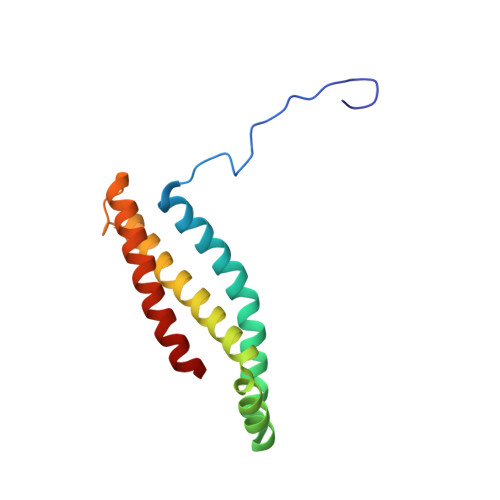
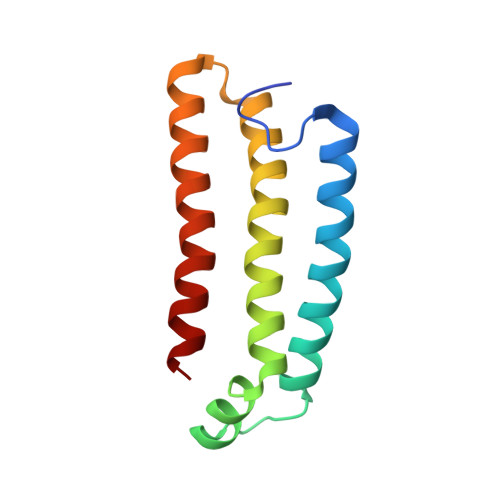
Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction.
Horsefield, R., Yankovskaya, V., Sexton, G., Whittingham, W., Shiomi, K., Omura, S., Byrne, B., Cecchini, G., Iwata, S.(2006) J Biological Chem 281: 7309-7316
- PubMed: 16407191
- DOI: https://doi.org/10.1074/jbc.M508173200
- Primary Citation of Related Structures:
2ACZ - PubMed Abstract:
The transfer of electrons and protons between membrane-bound respiratory complexes is facilitated by lipid-soluble redox-active quinone molecules (Q). This work presents a structural analysis of the quinone-binding site (Q-site) identified in succinate:ubiquinone oxidoreductase (SQR) from Escherichia coli. SQR, often referred to as Complex II or succinate dehydrogenase, is a functional member of the Krebs cycle and the aerobic respiratory chain and couples the oxidation of succinate to fumarate with the reduction of quinone to quinol (QH(2)). The interaction between ubiquinone and the Q-site of the protein appears to be mediated solely by hydrogen bonding between the O1 carbonyl group of the quinone and the side chain of a conserved tyrosine residue. In this work, SQR was co-crystallized with the ubiquinone binding-site inhibitor Atpenin A5 (AA5) to confirm the binding position of the inhibitor and reveal additional structural details of the Q-site. The electron density for AA5 was located within the same hydrophobic pocket as ubiquinone at, however, a different position within the pocket. AA5 was bound deeper into the site prompting further assessment using protein-ligand docking experiments in silico. The initial interpretation of the Q-site was re-evaluated in the light of the new SQR-AA5 structure and protein-ligand docking data. Two binding positions, the Q(1)-site and Q(2)-site, are proposed for the E. coli SQR quinone-binding site to explain these data. At the Q(2)-site, the side chains of a serine and histidine residue are suitably positioned to provide hydrogen bonding partners to the O4 carbonyl and methoxy groups of ubiquinone, respectively. This allows us to propose a mechanism for the reduction of ubiquinone during the catalytic turnover of the enzyme.
- Division of Molecular Biosciences, Imperial College London, London SW7 2AZ, United Kingdom.
Organizational Affiliation: