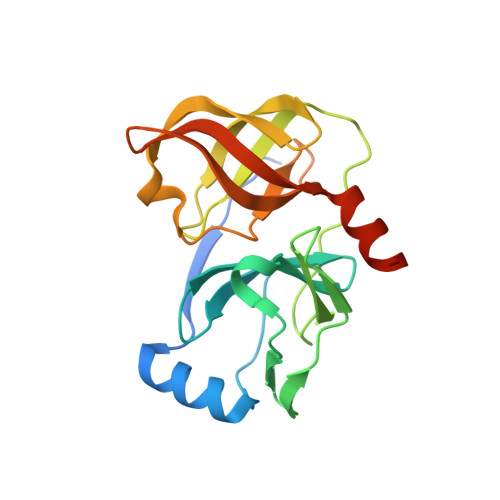
Hepatitis C Virus NS3-4A serine protease inhibitors: Use of a P2-P1 cyclopropyl alanine combination for improved potency.
Bogen, S., Saksena, A.K., Arasappan, A., Gu, H., Njoroge, F.G., Girijavallabhan, V., Pichardo, J., Butkiewicz, N., Prongay, A., Madison, V.(2005) Bioorg Med Chem Lett 15: 4515-4519
- PubMed: 16112862
- DOI: https://doi.org/10.1016/j.bmcl.2005.07.009
- Primary Citation of Related Structures:
2A4R - PubMed Abstract:
Modification of the P(2) and P(1) side chains of earlier P(3)-capped alpha-ketoamide inhibitor of HCV NS3 serine protease 1 resulted in the discovery of compound 24 with about 10-fold improvement in potency.
- Chemical Research, Schering Plough Research Institute, 2015 Galloping Hill Road, Kenilworth, NJ 07033, USA. stephane.bogen@spcorp.com
Organizational Affiliation: