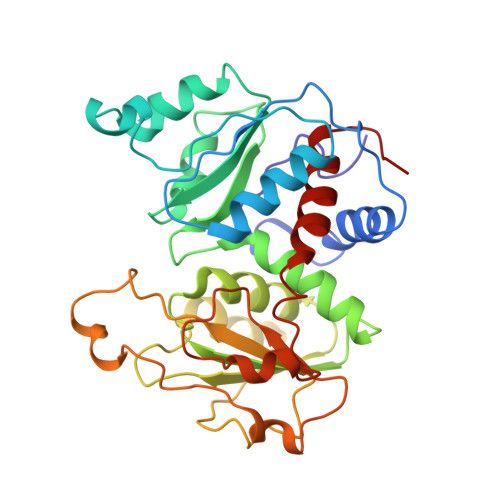
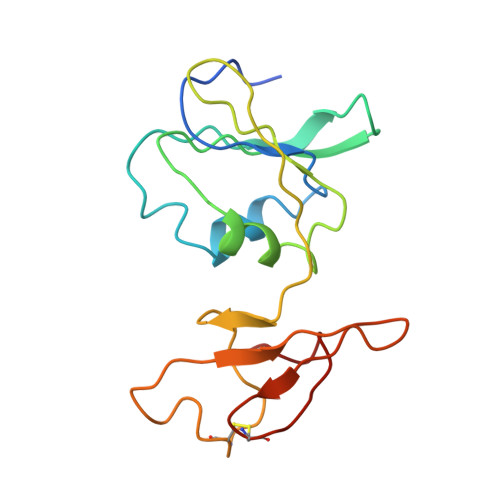
Structure of the E.coli Aspartate Transcarbamoylase Trapped in the Middle of the Catalytic Cycle.
Stieglitz, K.A., Dusinberre, K.J., Cardia, J.P., Tsuruta, H., Kantrowitz, E.R.(2005) J Mol Biology 352: 478-486
- PubMed: 16120448
- DOI: https://doi.org/10.1016/j.jmb.2005.07.046
- Primary Citation of Related Structures:
2A0F - PubMed Abstract:
Snapshots of the catalytic cycle of the allosteric enzyme aspartate transcarbamoylase have been obtained via X-ray crystallography. The enzyme in the high-activity high-affinity R state contains two catalytic chains in the asymmetric unit that are different. The active site in one chain is empty, while the active site in the other chain contains an analog of the first substrate to bind in the ordered mechanism of the reaction. Small angle X-ray scattering shows that once the enzyme is converted to the R state, by substrate binding, the enzyme remains in the R state until substrates are exhausted. Thus, this structure represents the active form of the enzyme trapped at two different stages in the catalytic cycle, before the substrates bind (or after the products are released), and after the first substrate binds. Opening and closing of the catalytic chain domains explains how the catalytic cycle occurs while the enzyme remains globally in the R-quaternary structure.
- Department of Chemistry, Merkert Chemistry Center, Boston College, Chestnut Hill, MA 02467, USA.
Organizational Affiliation: