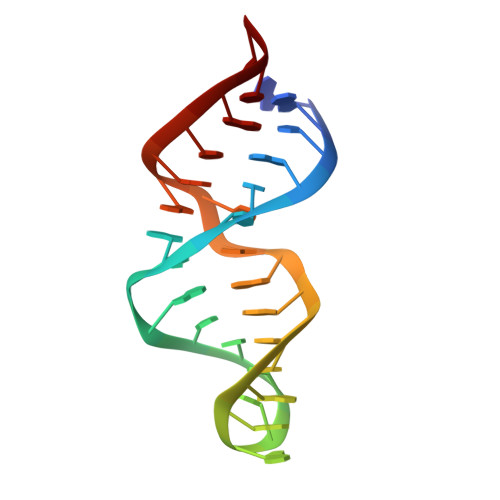
Structure of the most conserved internal loop in SRP RNA.
Schmitz, U., James, T.L., Lukavsky, P., Walter, P.(1999) Nat Struct Biol 6: 634-638
- PubMed: 10404218
- DOI: https://doi.org/10.1038/10683
- Primary Citation of Related Structures:
28SP, 28SR - PubMed Abstract:
The signal recognition particle (SRP) directs translating ribosomes to the protein translocation apparatus of endoplasmic reticulum (ER) membrane or the bacterial plasma membrane. The SRP is universally conserved, and in prokaryotes consists of two essential subunits, SRP RNA and SRP54, the latter of which binds to signal sequences on the nascent protein chains. Here we describe the solution NMR structure of a 28-mer RNA composing the most conserved part of SRP RNA to which SRP54 binds. Central to this function is a six-nucleotide internal loop that assumes a novel Mg2+-dependent structure with unusual cross-strand interactions; besides a cross-strand A/A stack, two guanines form hydrogen bonds with opposite-strand phosphates. The structure completely explains the phylogenetic conservation of the loop bases, underlining its importance for SRP54 binding and SRP function.
- Department of Pharmaceutical Chemistry, University of California San Francisco, 94143-0446, USA. schmitz@picasso.ucsf.edu
Organizational Affiliation: