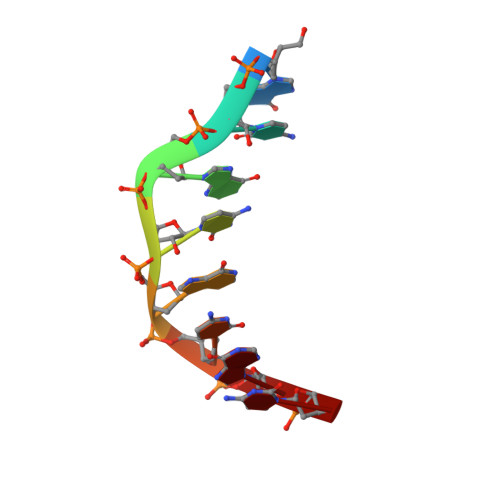
Crystal structures of B-form DNA-RNA chimers complexed with distamycin.
Chen, X., Ramakrishnan, B., Sundaralingam, M.(1995) Nat Struct Biol 2: 733-735
- PubMed: 7552741
- DOI: https://doi.org/10.1038/nsb0995-733
- Primary Citation of Related Structures:
216D, 217D - PubMed Abstract:
Two DNA-RNA chimers, complexed with DNA minor groove binding drugs, have been observed to adopt the B-form conformation for the first time. Thus, the RNA duplex may assume the B-DNA conformation when interacting with drugs, peptides or proteins.