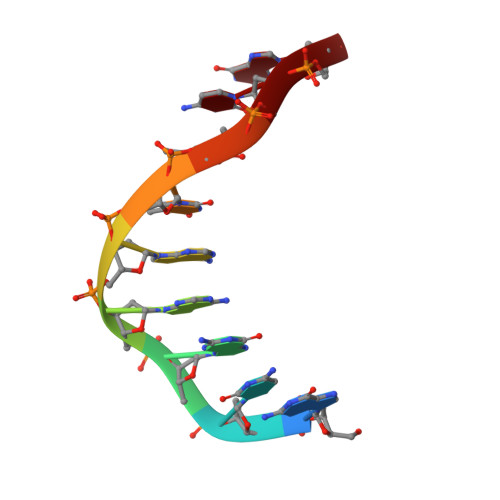
High-resolution structure of a DNA helix forming (C.G)*G base triplets.
Van Meervelt, L., Vlieghe, D., Dautant, A., Gallois, B., Precigoux, G., Kennard, O.(1995) Nature 374: 742-744
- PubMed: 7715732
- DOI: https://doi.org/10.1038/374742a0
- Primary Citation of Related Structures:
208D - PubMed Abstract:
Triple helices result from interaction between single- and double-stranded nucleic acids. Their formation is a possible mechanism for recombination of homologous gene sequences in nature and provides, inter alia, a basis for artificial control of gene activity. Triple-helix motifs have been extensively studied by a variety of techniques, but few high-resolution structural data are available. The only triplet structures characterized so far by X-ray diffraction were in protein-DNA complexes studied at about 3 A resolution. We report here the X-ray analysis of a DNA nonamer, d(GCGAATTCG), to a resolution of 2.05 A, in which the extended crystal structure contains (C.G)*G triplets as a fragment of triple helix. The guanosine-containing chains are in a parallel orientation. This arrangement is a necessary feature of models for homologous recombination which results ultimately in replacement of one length of DNA by another of similar sequence. The present-structure agrees with many published predictions of triplex organization, and provides an accurate representation of an element that allows sequence-specific association between single- and double-stranded nucleic acids.
- Department of Chemistry, Katholieke Universiteit Leuven, Heverlee, Belgium.
Organizational Affiliation: