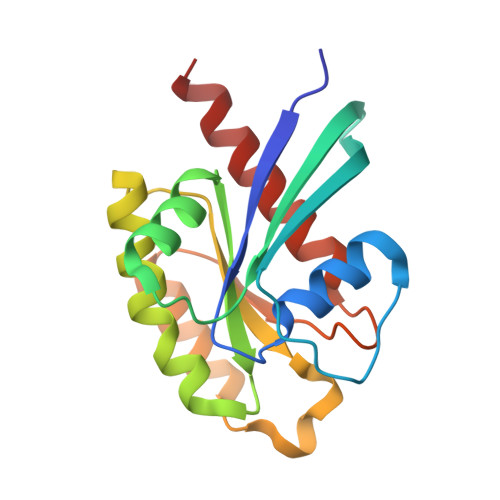
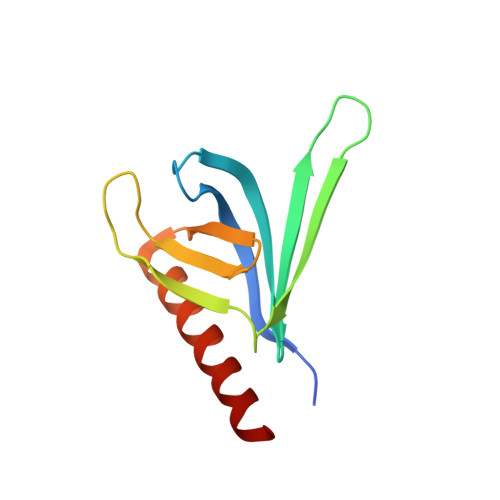
Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase.
Jin, R., Junutula, J.R., Matern, H.T., Ervin, K.E., Scheller, R.H., Brunger, A.T.(2005) EMBO J 24: 2064-2074
- PubMed: 15920473
- DOI: https://doi.org/10.1038/sj.emboj.7600699
- Primary Citation of Related Structures:
1ZC3, 1ZC4 - PubMed Abstract:
The Sec6/8 complex, also known as the exocyst complex, is an octameric protein complex that has been implicated in tethering of secretory vesicles to specific regions on the plasma membrane. Two subunits of the Sec6/8 complex, Exo84 and Sec5, have recently been shown to be effector targets for active Ral GTPases. However, the mechanism by which Ral proteins regulate the Sec6/8 activities remains unclear. Here, we present the crystal structure of the Ral-binding domain of Exo84 in complex with active RalA. The structure reveals that the Exo84 Ral-binding domain adopts a pleckstrin homology domain fold, and that RalA interacts with Exo84 via an extended interface that includes both switch regions. Key residues of Exo84 and RalA were found that determine the specificity of the complex interactions; these interactions were confirmed by mutagenesis binding studies. Structural and biochemical data show that Exo84 and Sec5 competitively bind to active RalA. Taken together, these results further strengthen the proposed role of RalA-regulated assembly of the Sec6/8 complex.
- Howard Hughes Medical Institute, Stanford University, Stanford, CA 94305-5432, USA.
Organizational Affiliation: