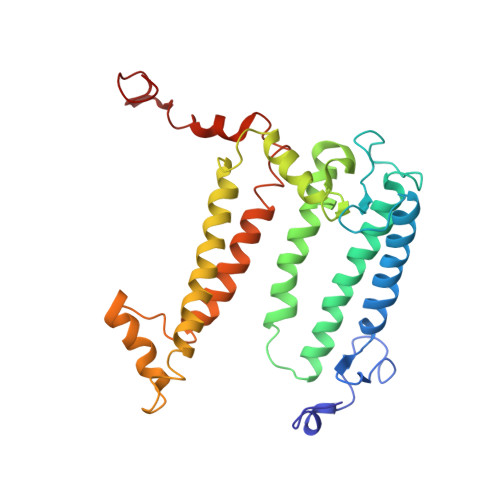
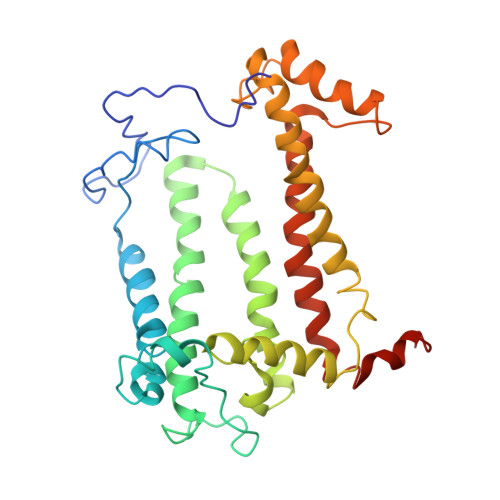
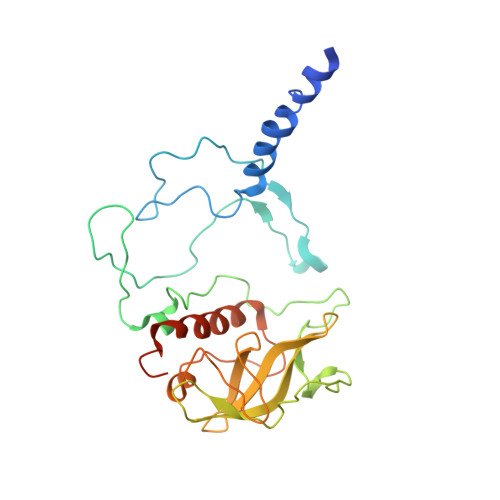
Design of a Redox-Linked Active Metal Site: Manganese Bound to Bacterial Reaction Centers at a Site Resembling That of Photosystem II
Uyeda, G., Camara-Artigas, A., Williams, J.C., Allen, J.P.(2005) Biochemistry 44: 7389-7394
- PubMed: 15895982
- DOI: https://doi.org/10.1021/bi050377n
- Primary Citation of Related Structures:
1Z9J, 1Z9K - PubMed Abstract:
Metals bound to proteins perform a number of crucial biological reactions, including the oxidation of water by a manganese cluster in photosystem II. Although evolutionarily related to photosystem II, bacterial reaction centers lack both a strong oxidant and a manganese cluster for mediating the multielectron and proton transfer needed for water oxidation. In this study, carboxylate residues were introduced by mutagenesis into highly oxidizing reaction centers at a site homologous to the manganese-binding site of photosystem II. In the presence of manganese, light-minus-dark difference optical spectra of reaction centers from the mutants showed a lack of the oxidized bacteriochlorophyll dimer, while the reduced primary quinone was still present, demonstrating that manganese was serving as a secondary electron donor. On the basis of these steady-state optical measurements, the mutant with the highest-affinity site had a dissociation constant of approximately 1 microM. For the highest-affinity mutant, a first-order rate with a lifetime of 12 ms was observed for the reduction of the oxidized bacteriochlorophyll dimer by the bound manganese upon exposure to light. The dependence of the amplitude of this component on manganese concentration yielded a dissociation constant of approximately 1 muM, similar to that observed in the steady-state measurements. The three-dimensional structure determined by X-ray diffraction of the mutant with the high-affinity site showed that the binding site contains a single bound manganese ion, three carboxylate groups (including two groups introduced by mutagenesis), a histidine residue, and a bound water molecule. These reaction centers illustrate the successful design of a redox active metal center in a protein complex.
- Department of Chemistry and Biochemistry, Arizona State University, Tempe, Arizona 85287, USA.
Organizational Affiliation: