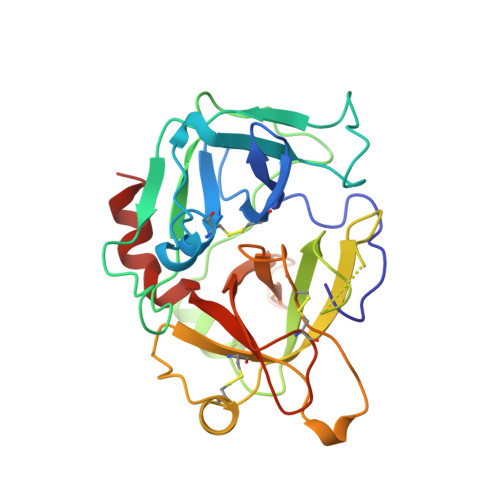
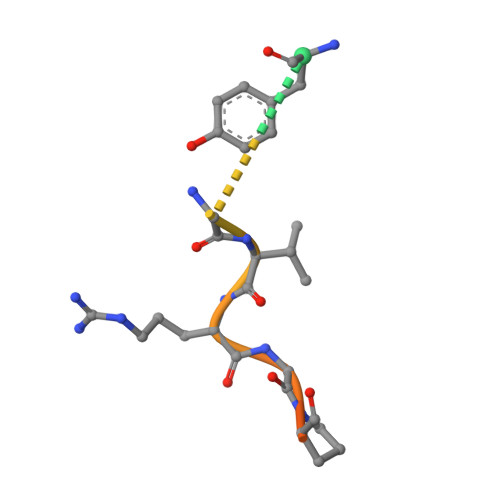
Crystal structure of fibrinogen-Aalpha peptide 1-23 (F8Y) bound to bovine thrombin explains why the mutation of Phe-8 to tyrosine strongly inhibits normal cleavage at Arg-16.
Malkowski, M.G., Martin, P.D., Lord, S.T., Edwards, B.F.(1997) Biochem J 326: 815-822
- PubMed: 9307032
- DOI: https://doi.org/10.1042/bj3260815
- Primary Citation of Related Structures:
1YCP - PubMed Abstract:
A peptide containing residues 1-50 of the Aalpha-chain of fibrinogen, expressed as a fusion peptide with beta-galactosidase, is rapidly cleaved by thrombin at Arg-16, similarly to whole fibrinogen. When Phe-8, which is highly conserved, is replaced with tyrosine (F8Y), the cleavage is slowed drastically [Lord, Byrd, Hede, Wei and Colby (1990) J. Biol. Chem. 265, 838-843]. To examine the structural basis for this result, we have determined the crystal structure of bovine thrombin complexed with a synthetic peptide containing residues 1-23 of fibrinogen Aalpha and the F8Y mutation. The crystals are in space group P43212, with unit-cell dimensions of a = 88.3 A (1 A = 0.1 nm), c = 195.5 A and two complexes in the asymmetric unit. The final R factor is 0.183 for 2sigma data from 7.0 to 2.5 A resolution. There is continuous density for the five residues in the P3, P2, P1, P1' and P2' positions of the peptide (Gly-14f to Pro-18f) at the active site of thrombin, and isolated but well-defined density for Tyr-8f at position P9 in the hydrophobic pocket of thrombin. The tyrosine residue is shifted relative to phenylalanine in the native peptide because the phenol side chain is larger and makes a novel, intrapeptide hydrogen bond with Gly-14f. Adjacent peptide residues cannot form the hydrogen bonds that stabilize the secondary structure of the native peptide. Consequently, the 'reaction'geometry at the scissile bond, eight residues from the mutation, is perturbed and the peptide is mostly uncleaved in the crystal structure.
- Department of Biochemistry, Wayne State University, 540 E. Canfield Avenue, Detroit, MI 48201, USA.
Organizational Affiliation: