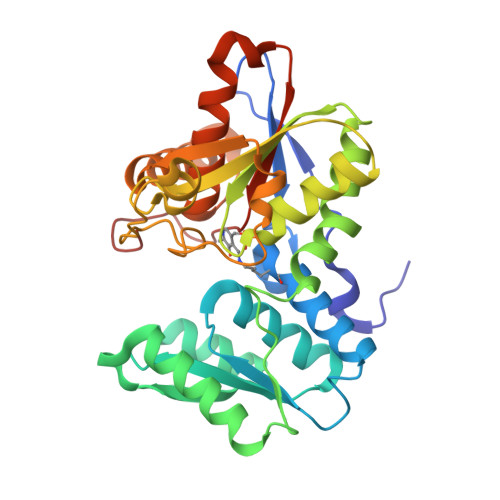
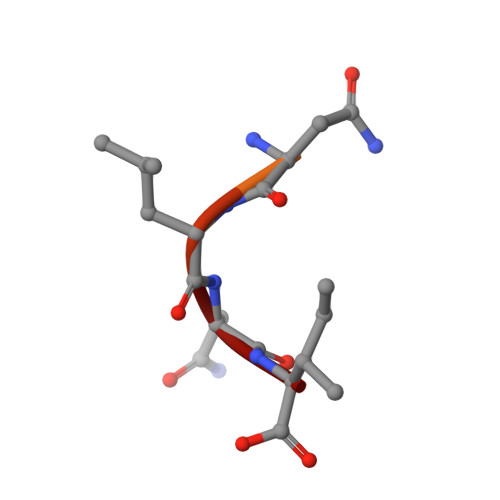
The active site of O-acetylserine sulfhydrylase is the anchor point for bienzyme complex formation with serine acetyltransferase.
Huang, B., Vetting, M.W., Roderick, S.L.(2005) J Bacteriol 187: 3201-3205
- PubMed: 15838047
- DOI: https://doi.org/10.1128/JB.187.9.3201-3205.2005
- Primary Citation of Related Structures:
1Y7L - PubMed Abstract:
The biosynthesis of cysteine in bacteria and plants is carried out by a two-step pathway, catalyzed by serine acetyltransferase (SAT) and O-acetylserine sulfhydrylase (OASS; O-acetylserine [thiol] lyase). The aerobic form of OASS forms a tight bienzyme complex with SAT in vivo, termed cysteine synthase. We have determined the crystal structure of OASS in complex with a C-terminal peptide of SAT required for bienzyme complex formation. The binding site of the peptide is at the active site of OASS, and its C-terminal carboxyl group occupies the same anion binding pocket as the alpha-carboxylate of the O-acetylserine substrate of OASS. These results explain the partial inhibition of OASS by SAT on complex formation as well as the competitive dissociation of the complex by O-acetylserine.
- Department of Biochemistry, Albert Einstein College of Medicine, 1300 Morris Park Ave., Bronx, NY 10461, USA.
Organizational Affiliation: