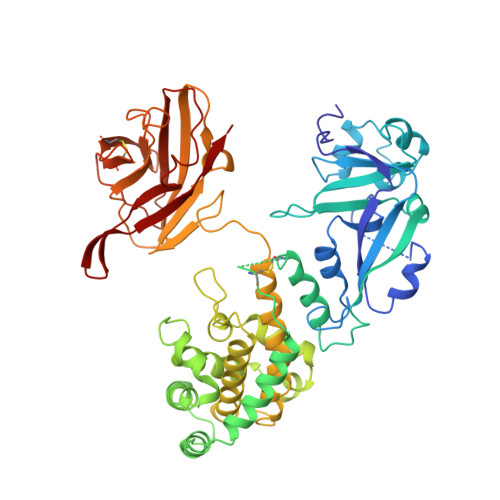
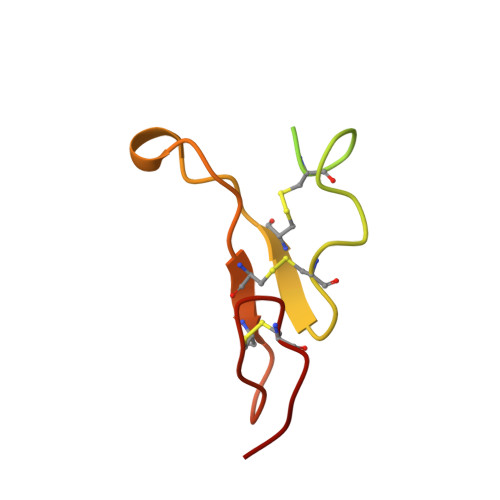
Crystal structure of the complex of diphtheria toxin with an extracellular fragment of its receptor.
Louie, G.V., Yang, W., Bowman, M.E., Choe, S.(1997) Mol Cell 1: 67-78
- PubMed: 9659904
- DOI: https://doi.org/10.1016/s1097-2765(00)80008-8
- Primary Citation of Related Structures:
1XDT - PubMed Abstract:
We describe the crystal structure at 2.65 A resolution of diphtheria toxin (DT) complexed 1:1 with a fragment of its cell-surface receptor, the precursor of heparin-binding epidermal-growth-factor-like growth factor (HBEGF). HBEGF in the complex has the typical EGF-like fold and packs its principal beta hairpin against the face of a beta sheet in the receptor-binding domain of DT. The interface has a predominantly hydrophobic core, and polar interactions are formed at the periphery. The structure of the complex suggests that part of the membrane anchor of the receptor can interact with a hinge region of DT. The toxin molecule is thereby induced to form an open conformation conducive to membrane insertion. The structure provides a basis for altering the binding specificity of the toxin, and may also serve as a model for other EGF-receptor interactions.
- Structural Biology Laboratory, Salk Institute for Biological Studies, La Jolla, California 92037, USA.
Organizational Affiliation: