Crystal Structure of Recbcd Enzyme Reveals a Machine for Processing DNA Breaks
Singleton, M.R., Dillingham, M.S., Gaudier, M., Kowalczykowski, S.C., Wigley, D.B.(2004) Nature 432: 187
- PubMed: 15538360
- DOI: https://doi.org/10.1038/nature02988
- Primary Citation of Related Structures:
1W36 - PubMed Abstract:
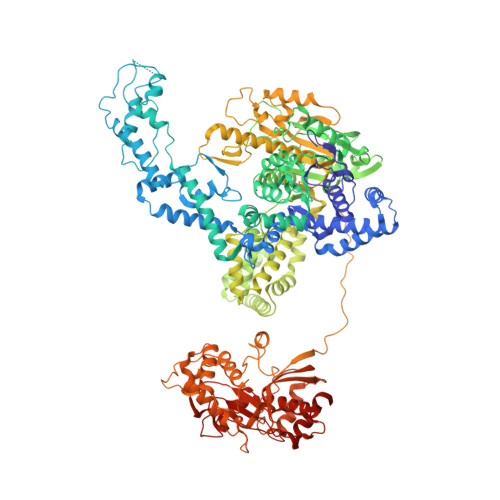
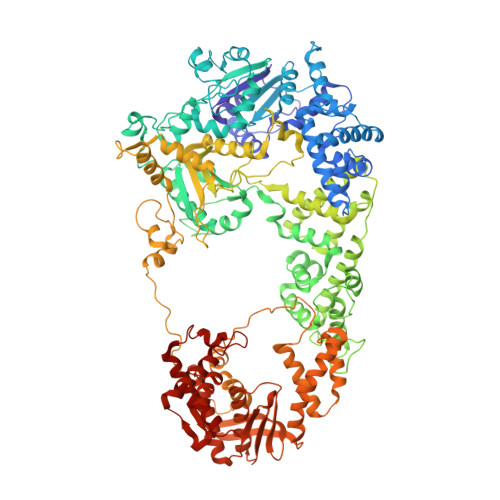
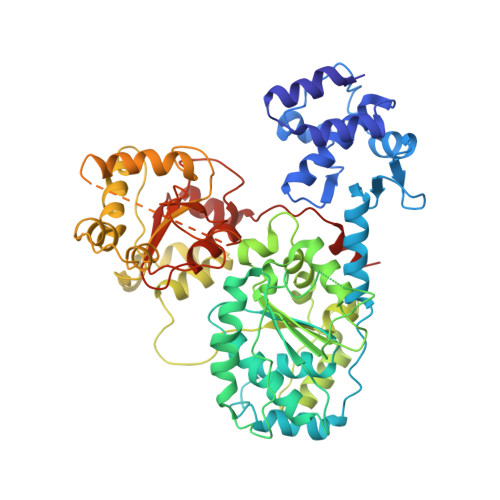
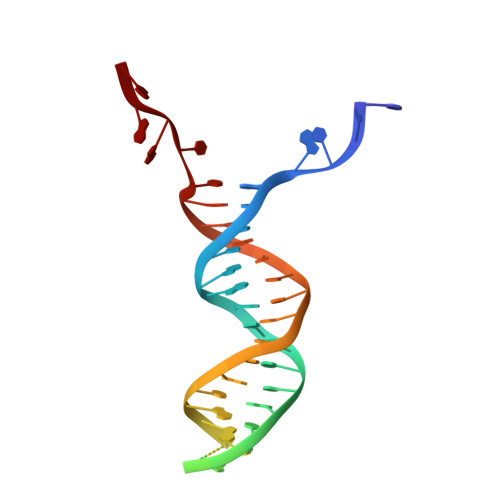
RecBCD is a multi-functional enzyme complex that processes DNA ends resulting from a double-strand break. RecBCD is a bipolar helicase that splits the duplex into its component strands and digests them until encountering a recombinational hotspot (Chi site). The nuclease activity is then attenuated and RecBCD loads RecA onto the 3' tail of the DNA. Here we present the crystal structure of RecBCD bound to a DNA substrate. In this initiation complex, the DNA duplex has been split across the RecC subunit to create a fork with the separated strands each heading towards different helicase motor subunits. The strands pass along tunnels within the complex, both emerging adjacent to the nuclease domain of RecB. Passage of the 3' tail through one of these tunnels provides a mechanism for the recognition of a Chi sequence by RecC within the context of double-stranded DNA. Gating of this tunnel suggests how nuclease activity might be regulated.
- Cancer Research UK Clare Hall Laboratories, The London Research Institute, Blanche Lane, South Mimms, Potters Bar, Herts. EN6 3LD, UK.
Organizational Affiliation: