Conformational Changes in the Phosphorylated C-Terminal Domain of Rhodopsin During Rhodopsin Arrestin Interactions
Kisselev, O.G., Downs, M.A., Mcdowell, J.H., Hargrave, P.A.(2004) J Biological Chem 279: 51203
- PubMed: 15351781
- DOI: https://doi.org/10.1074/jbc.M407341200
- Primary Citation of Related Structures:
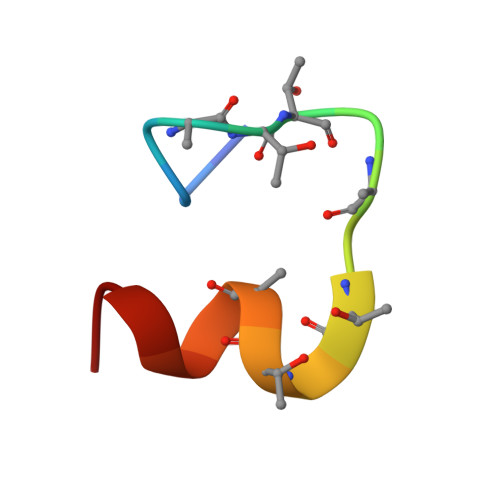
1VQX - PubMed Abstract:
Phosphorylation of activated G-protein-coupled receptors and the subsequent binding of arrestin mark major molecular events of homologous desensitization. In the visual system, interactions between arrestin and the phosphorylated rhodopsin are pivotal for proper termination of visual signals. By using high resolution proton nuclear magnetic resonance spectroscopy of the phosphorylated C terminus of rhodopsin, represented by a synthetic 7-phosphopolypeptide, we show that the arrestin-bound conformation is a well ordered helix-loop structure connected to rhodopsin via a flexible linker. In a model of the rhodopsin-arrestin complex, the phosphates point in the direction of arrestin and form a continuous negatively charged surface, which is stabilized by a number of positively charged lysine and arginine residues of arrestin. Opposite to the mostly extended structure of the unphosphorylated C-terminal domain of rhodopsin, the arrestin-bound C-terminal helix is a compact domain that occupies a central position between the cytoplasmic loops and occludes the key binding sites of transducin. In conjunction with other binding sites, the helix-loop structure provides a mechanism of shielding phosphates in the center of the rhodopsin-arrestin complex and appears critical in guiding arrestin for high affinity binding with rhodopsin.
- Department of Ophthalmology, St. Louis University School of Medicine, St. Louis, Missouri 63104, USA. kisselev@slu.edu
Organizational Affiliation: