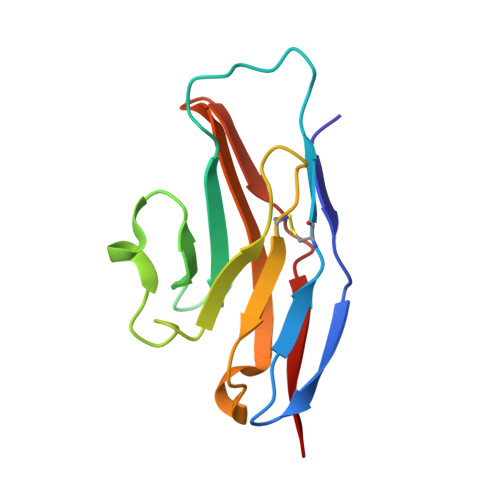
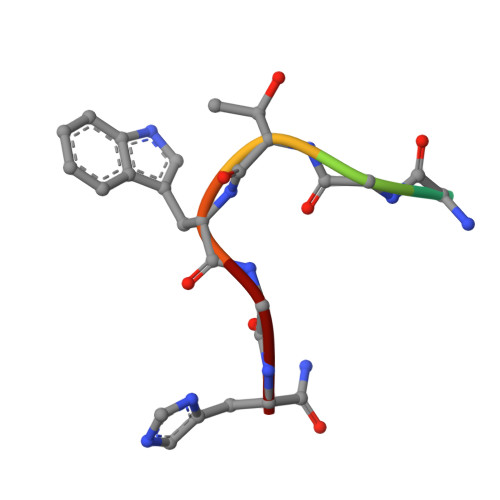
Complex of Sialoadhesin with a Glycopeptide Ligand
Bukrinsky, J.T., Hilaire, P.M.S., Meldal, M., Crocker, P.R., Henriksen, A.(2004) Biochim Biophys Acta 1702: 173
- PubMed: 15488769
- DOI: https://doi.org/10.1016/j.bbapap.2004.08.015
- Primary Citation of Related Structures:
1URL - PubMed Abstract:
Sialoadhesin is a sialic acid-binding immunoglobulin-like lectin (Siglec), expressed on subsets of macrophages. It is a model system for Siglec receptor-mediated cell surface interactions through binding of sialylated glycoconjugates. The N-terminal sialoadhesin domain can mediate sialic acid-binding on its own. The structure of this domain has been determined in complex with a sialic acid-containing heptapeptide, (Ala-Gly-His-Thr(Neu5Ac)-Trp-Gly-His). The affinity of sialoadhesin for this ligand is four times higher than the affinity for the natural linkage 2,3'-sialyllactose. The structure of the glycopeptide complex suggests strategies for ligand optimization and provides possible explanations for the observed differences in specificities among the Siglecs.
- Department of Chemistry, The Carlsberg Laboratory, Gamle Carlsberg Vej 10, DK-2500 Valby, Denmark.
Organizational Affiliation: