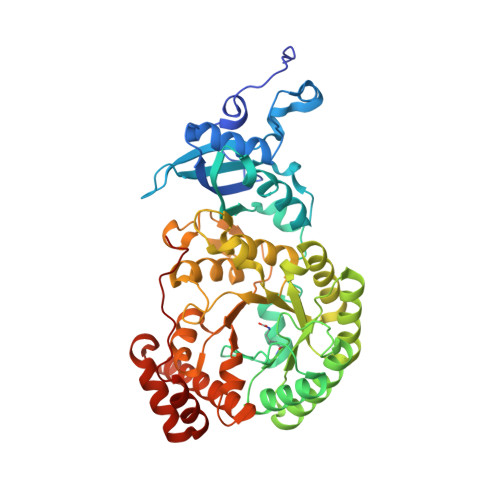
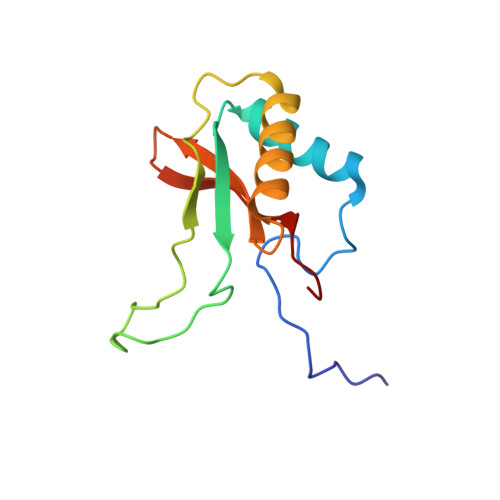
Calcium Supports Loop Closure But not Catalysis in Rubisco
Karehabadi, S., Taylor, T.C., Andersson, I.(2003) J Mol Biology 334: 65
- PubMed: 14596800
- DOI: https://doi.org/10.1016/j.jmb.2003.09.025
- Primary Citation of Related Structures:
1UPM, 1UPP - PubMed Abstract:
Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase) catalyses CO(2) assimilation in biology. A prerequisite for catalysis is an activation process, whereby an active site lysine is selectively carbamylated. The carbamyl group is then stablised by a metal ion, which in vivo is Mg(2+). Other divalent metal ions can replace Mg(2+) as activators in vitro, but the nature of the metal ion strongly influences the catalytic activity of the enzyme and has a differential effect on the ratio of the carboxylation reaction and the competing oxygenation reaction. Biochemical studies show that calcium promotes carbamylation but not catalysis. To investigate the role of the metal in catalysis, we have determined two structures of the enzyme complexed with Ca(2+) and the transition state analogue 2-carboxy-D-arbinitol-1,5-bisphosphate (2CABP). One of the complexes was prepared by soaking 2CABP into crystals of the enzyme-Ca(2+)-product complex, while the other was obtained by cocrystallising the enzyme with calcium and 2CABP under activating conditions. The two crystals belong to different space groups, and one was merohedrally twinned. Both complexes show very similar three-dimensional features. The enzyme is carbamylated at Lys201, and requisite loops close over the bound ligands in the active site, shielding them from the solvent in a manner similar to the corresponding complex with Mg(2+). However, there are subtle differences that could explain the particular role of Ca(2+) in these processes. The larger radius of the calcium ion and its reduced Lewis-acid character causes a significant increase in the required proton hop distance between the C3 proton and the carbamate on Lys201 in the calcium complex. This alone could explain the inability of calcium to sustain catalysis in Rubisco. Similar effects are also expected on subsequent proton transfer steps in the catalytic cycle. Here we also discuss the effect of metal substitution on the dynamics of the ligands around the metal ion.
- Department of Molecular Biosciences, Swedish University of Agricultural Sciences, BMC Box 590, S-751 24, Uppsala, Sweden.
Organizational Affiliation: