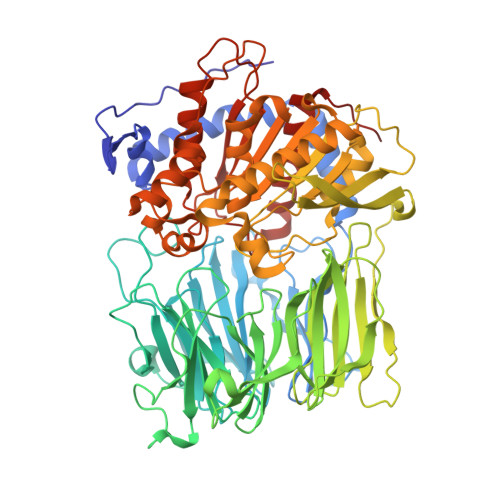
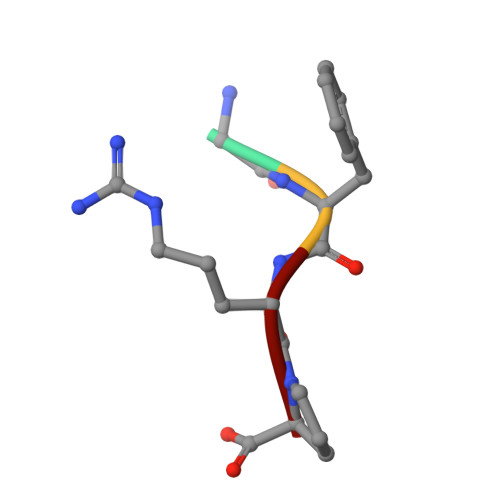
Electrostatic Environment at the Active Site of Prolyl Oligopeptidase is Highly Influential During Substrate Binding
Szeltner, Z., Rea, D., Renner, V., Juliano, L., Fulop, V., Polgar, L.(2003) J Biological Chem 278: 48786
- PubMed: 14514675
- DOI: https://doi.org/10.1074/jbc.M309555200
- Primary Citation of Related Structures:
1UOO, 1UOP, 1UOQ - PubMed Abstract:
The positive electrostatic environment of the active site of prolyl oligopeptidase was investigated by using substrates with glutamic acid at positions P2, P3, P4, and P5, respectively. The different substrates gave various pH rate profiles. The pKa values extracted from the curves are apparent parameters, presumably affected by the nearby charged residues, and do not reflect the ionization of a simple catalytic histidine as found in the classic serine peptidases like chymotrypsin and subtilisin. The temperature dependence of kcat/Km did not produce linear Arrhenius plots, indicating different changes in the individual rate constants with the increase in temperature. This rendered it possible to calculate these constants, i.e. the formation (k1) and decomposition (k-1) of the enzyme-substrate complex and the acylation constant (k2), as well as the corresponding activation energies. The results have revealed the relationship between the complex Michaelis parameters and the individual rate constants. Structure determination of the enzyme-substrate complexes has shown that the different substrates display a uniform binding mode. None of the glutamic acids interacts with a charged group. We conclude that the specific rate constant is controlled by k1 rather than k2 and that the charged residues from the substrate and the enzyme can markedly affect the formation but not the structure of the enzyme-substrate complexes.
- Institute of Enzymology, Biological Research Center, Hungarian Academy of Sciences, PO Box 7, H-1518 Budapest 112, Hungary.
Organizational Affiliation: