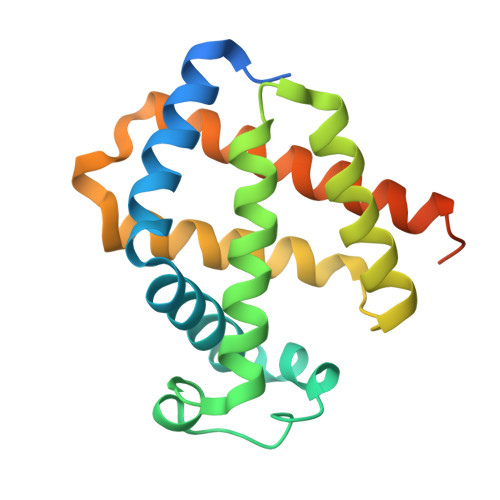
Crystal structure of cytoglobin: the fourth globin type discovered in man displays heme hexa-coordination.
de Sanctis, D., Dewilde, S., Pesce, A., Moens, L., Ascenzi, P., Hankeln, T., Burmester, T., Bolognesi, M.(2004) J Mol Biology 336: 917-927
- PubMed: 15095869
- DOI: https://doi.org/10.1016/j.jmb.2003.12.063
- Primary Citation of Related Structures:
1UMO, 1URV, 1UT0 - PubMed Abstract:
Cytoglobin is a recently discovered hemeprotein belonging to the globin superfamily together with hemoglobin, myoglobin and neuroglobin. Although distributed in almost all human tissues, cytoglobin has not been ascribed a specific function. Human cytoglobin is composed of 190 amino acid residues. Sequence alignments show that a protein core region (about 150 residues) is structurally related to hemoglobin and myoglobin, being complemented by about 20 extra residues both on the N and C termini. In the absence of exogenous ligands (e.g. O2), the cytoglobin distal HisE7 residue is coordinated to the heme Fe atom, thus decreasing the ligand affinity. The crystal structure of human cytoglobin (2.1 A resolution, 21.3% R-factor) highlights a three-over-three alpha-helical globin fold, covering residues 18-171; the 1-17 N-terminal, and the 172-190 C-terminal residue segments are disordered in both molecules of the crystal asymmetric unit. Heme hexa-coordination is evident in one of the two cytoglobin chains, whereas alternate conformation for the heme distal region, achieving partial heme penta-coordination, is observed in the other. Human cytoglobin displays a large apolar protein matrix cavity, next to the heme, not related to the myoglobin cavities recognized as temporary ligand docking stations. The cavity, which may provide a heme ligand diffusion pathway, is connected to the external space through a narrow tunnel nestled between the globin G and H helices.
- Department of Physics-INFM, Centre for Excellence in Biomedical Research, University of Genova, Via Dodecaneso 33, 1-16146 Genova, Italy.
Organizational Affiliation: