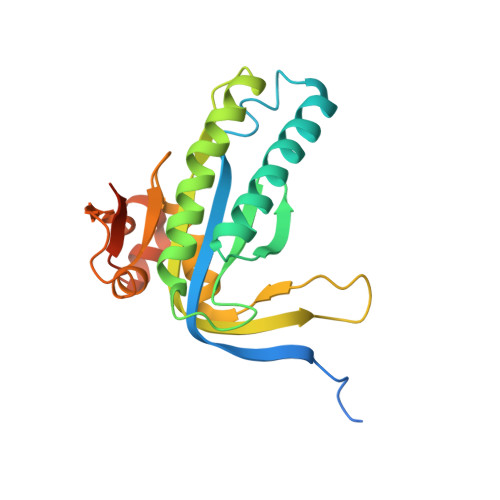
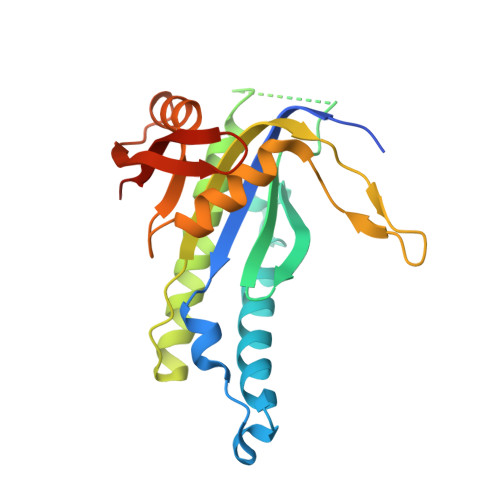
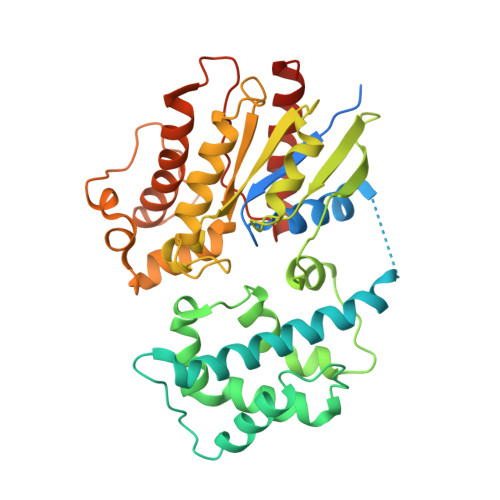
Structural basis for the inhibition of mammalian membrane adenylyl cyclase by 2 '(3')-O-(N-Methylanthraniloyl)-guanosine 5 '-triphosphate.
Mou, T.C., Gille, A., Fancy, D.A., Seifert, R., Sprang, S.R.(2005) J Biological Chem 280: 7253-7261
- PubMed: 15591060
- DOI: https://doi.org/10.1074/jbc.M409076200
- Primary Citation of Related Structures:
1TL7, 1U0H - PubMed Abstract:
Membrane-bound mammalian adenylyl cyclase (mAC) catalyzes the synthesis of intracellular cyclic AMP from ATP and is activated by stimulatory G protein alpha subunits (Galpha(s)) and by forskolin (FSK). mACs are inhibited with high potency by 2 '(3')-O-(N-methylanthraniloyl) (MANT)-substituted nucleotides. In this study, the crystal structures of the complex between Galpha(s).GTPgammaS and the catalytic C1 and C2 domains from type V and type II mAC (VC1.IIC2), bound to FSK and either MANT-GTP.Mg(2+) or MANT-GTP.Mn(2+) have been determined. MANT-GTP coordinates two metal ions and occupies the same position in the catalytic site as P-site inhibitors and substrate analogs. However, the orientation of the guanine ring is reversed relative to that of the adenine ring. The MANT fluorophore resides in a hydrophobic pocket at the interface between the VC1 and IIC2 domains and prevents mAC from undergoing the "open" to "closed" domain rearrangement. The K(i) of MANT-GTP for inhibition of VC1.IIC2 is lower in the presence of mAC activators and lower in the presence of Mn(2+) compared with Mg(2+), indicating that the inhibitor binds more tightly to the catalytically most active form of the enzyme. Fluorescence resonance energy transfer-stimulated emission from the MANT fluorophore upon excitation of Trp-1020 in the MANT-binding pocket of IIC2 is also stronger in the presence of FSK. Mutational analysis of two non-conserved amino acids in the MANT-binding pocket suggests that residues outside of the binding site influence isoform selectivity toward MANT-GTP.
- Department of Biochemistry, Howard Hughes Medical Institute, The University of Texas Southwestern Medical, Dallas, Texas 75390-9050, USA.
Organizational Affiliation: