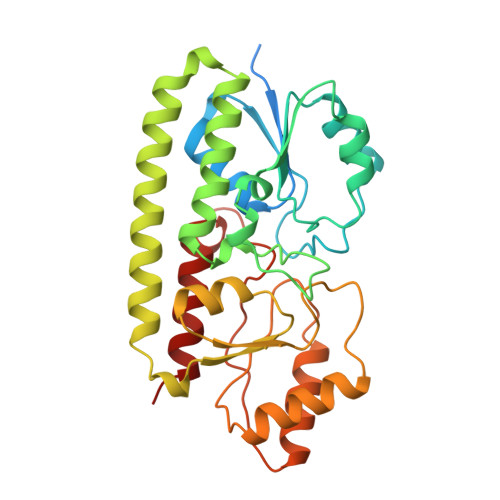
Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone.
Lee, Y.H., Deka, R.K., Norgard, M.V., Radolf, J.D., Hasemann, C.A.(1999) Nat Struct Biol 6: 628-633
- PubMed: 10404217
- DOI: https://doi.org/10.1038/10677
- Primary Citation of Related Structures:
1TOA - PubMed Abstract:
The crystal structure of recombinant TroA, a zinc-binding protein component of an ATP-binding cassette transport system in Treponema pallidum, was determined at a resolution of 1.8 A. The organization of the protein is largely similar to other periplasmic ligand-binding proteins (PLBP), in that two independent globular domains interact with each other to create a zinc-binding cleft between them. The structure has one bound zinc pentavalently coordinated to residues from both domains. Unlike previous PLBP structures that have an interdomain hinge composed of beta-strands, the N- and C-domains of TroA are linked by a single long backbone helix. This unique backbone helical conformation was possibly adopted to limit the hinge motion associated with ligand exchange.
- Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas 75235-8884, USA.
Organizational Affiliation: