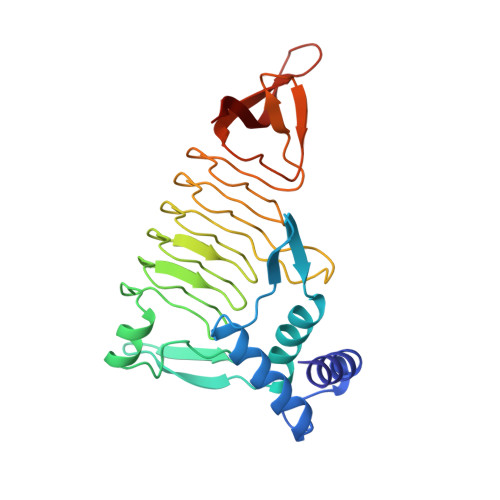
Three-dimensional structure of tetrahydrodipicolinate N-succinyltransferase.
Beaman, T.W., Binder, D.A., Blanchard, J.S., Roderick, S.L.(1997) Biochemistry 36: 489-494
- PubMed: 9012664
- DOI: https://doi.org/10.1021/bi962522q
- Primary Citation of Related Structures:
1TDT - PubMed Abstract:
The conversion of tetrahydrodipicolinate and succinyl-CoA to N-succinyltetrahydrodipicolinate and CoA is catalyzed by tetrahydrodipicolinate N-succinyltransferase and is the committed step in the succinylase pathway by which bacteria synthesize L-lysine and meso-diaminopimelate, a component of peptidoglycan. The X-ray crystal structure of THDP succinyltransferase has been determined to 2.2 A resolution and has been refined to a crystallographic R-factor of 17.0%. The enzyme is trimeric and displays the left-handed parallel beta-helix (L beta H) structural motif encoded by the "hexapeptide repeat" amino acid sequence motif [Raetz, C.R.H., & Roderick, S.L. (1995) Science 270, 997-1000]. The approximate location of the active site of THDP succinyltransferase is suggested by the proximity of binding sites for two inhibitors: p-(chloromercuri)benzenesulfonic acid and cobalt ion, both of which bind to the L beta H domain.
- Department of Biochemistry, Albert Einstein College of Medicine, Bronx, New York 10461, USA.
Organizational Affiliation: