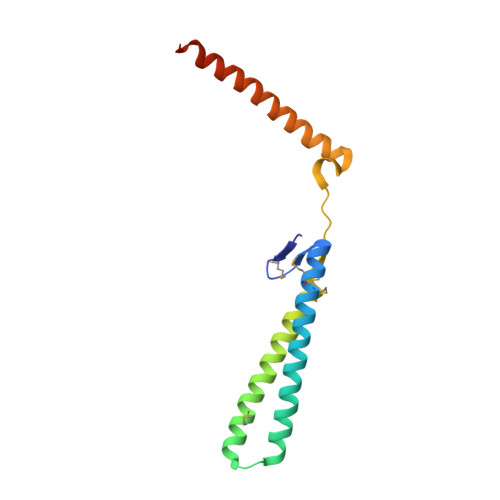
Structural genomics of Caenorhabditis elegans: structure of the BAG domain.
Symersky, J., Zhang, Y., Schormann, N., Li, S., Bunzel, R., Pruett, P., Luan, C.H., Luo, M.(2004) Acta Crystallogr D Biol Crystallogr 60: 1606-1610
- PubMed: 15333932
- DOI: https://doi.org/10.1107/S0907444904017603
- Primary Citation of Related Structures:
1T7S - PubMed Abstract:
Binding of the BAG domain to the eukaryotic chaperone heat-shock protein (Hsp70) promotes ATP-dependent release of the protein substrate from Hsp70. Although the murine and human BAG domains have been shown to form an antiparallel three-helix bundle, the Caenorhabditis elegans BAG domain is formed by two antiparallel helices, while the third helix is extended away and stabilized by crystal-packing interactions. A small beta-sheet between helices 2 and 3 interferes with formation of the intramolecular three-helix bundle. However, intermolecular three-helix bundles are observed throughout the crystal packing and suggest that stable functional dimers and tetramers can be formed in solution. The structure may represent a new folding type of the BAG domain.
- Southeast Collaboratory for Structural Genomics, Center for Biophysical Sciences and Engineering, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
Organizational Affiliation: