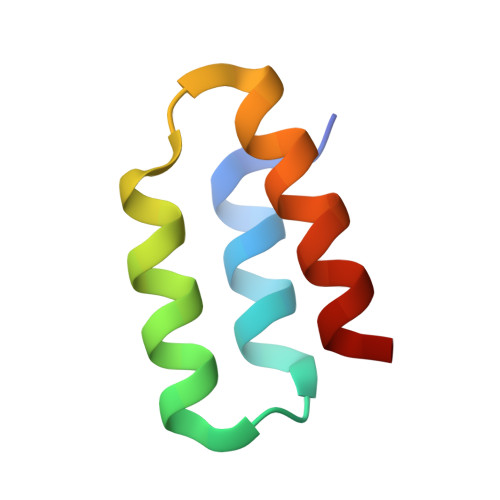
Structural basis for the attachment of a paramyxoviral polymerase to its template.
Kingston, R.L., Hamel, D.J., Gay, L.S., Dahlquist, F.W., Matthews, B.W.(2004) Proc Natl Acad Sci U S A 101: 8301-8306
- PubMed: 15159535
- DOI: https://doi.org/10.1073/pnas.0402690101
- Primary Citation of Related Structures:
1T6O - PubMed Abstract:
The nucleocapsid of measles virus is the template for viral RNA synthesis and is generated through packaging of the genomic RNA by the nucleocapsid protein (N). The viral polymerase associates with the nucleocapsid through a small, trihelical binding domain at the carboxyl terminus of the phosphoprotein (P). Translocation of the polymerase along the nucleocapsid during RNA synthesis is thought to involve the repeated attachment and release of the binding domain. We have investigated the interaction between the binding domain from measles P (amino acids 457-507) and the sequence it recognizes within measles N (amino acids 477-505). By using both solution NMR spectroscopy and x-ray crystallography, we show that N(487-503) binds as a helix to the surface created by the second (alpha2) and third (alpha3) helices of P(457-507), in an orientation parallel to the helix alpha3, creating a four-helix bundle. The binding interface is tightly packed and dominated by hydrophobic amino acids. Binding and folding of N(487-503) are coupled. However, when not bound to P, N(487-503) does not resemble a statistical random coil but instead exists in a loosely structured state that mimics the bound conformation. We propose that before diffusional encounter, the ensemble of accessible conformations for N(487-503) is biased toward structures capable of binding P, facilitating rapid association of the two proteins. This study provides a structural analysis of polymerase-template interactions in a paramyxovirus and presents an example of a protein-protein interaction that must be only transiently maintained as part of its normal function.
- Howard Hughes Medical Institute, Institute of Molecular Biology, University of Oregon, Eugene, OR 97403, USA. richard@uoxray.uoregon.edu
Organizational Affiliation: