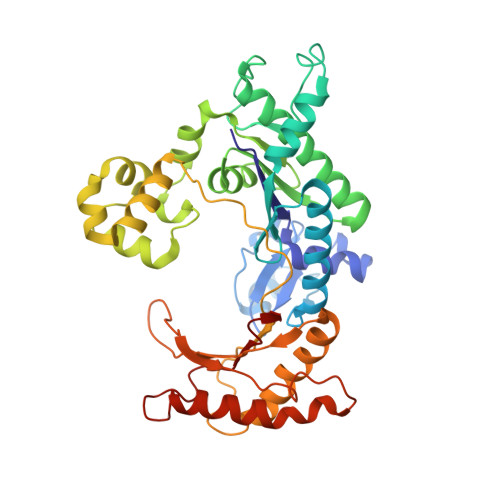
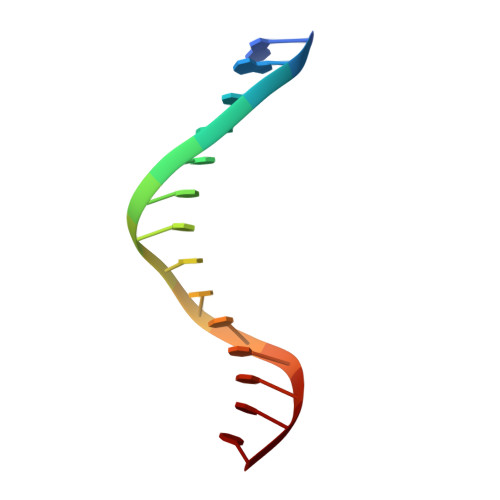
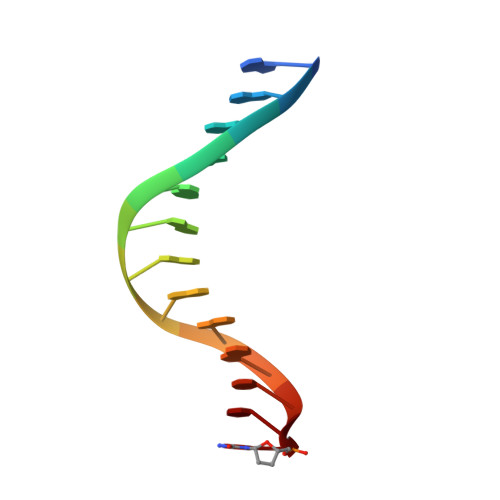
Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing.
Nair, D.T., Johnson, R.E., Prakash, S., Prakash, L., Aggarwal, A.K.(2004) Nature 430: 377-380
- PubMed: 15254543
- DOI: https://doi.org/10.1038/nature02692
- Primary Citation of Related Structures:
1T3N - PubMed Abstract:
Almost all DNA polymerases show a strong preference for incorporating the nucleotide that forms the correct Watson-Crick base pair with the template base. In addition, the catalytic efficiencies with which any given polymerase forms the four possible correct base pairs are roughly the same. Human DNA polymerase-iota (hPoliota), a member of the Y family of DNA polymerases, is an exception to these rules. hPoliota incorporates the correct nucleotide opposite a template adenine with a several hundred to several thousand fold greater efficiency than it incorporates the correct nucleotide opposite a template thymine, whereas its efficiency for correct nucleotide incorporation opposite a template guanine or cytosine is intermediate between these two extremes. Here we present the crystal structure of hPoliota bound to a template primer and an incoming nucleotide. The structure reveals a polymerase that is 'specialized' for Hoogsteen base-pairing, whereby the templating base is driven to the syn conformation. Hoogsteen base-pairing offers a basis for the varied efficiencies and fidelities of hPoliota opposite different template bases, and it provides an elegant mechanism for promoting replication through minor-groove purine adducts that interfere with replication.
- Structural Biology Program, Department of Physiology and Biophysics, Mount Sinai School of Medicine, Box 1677, 1425 Madison Avenue, New York, New York 10029, USA.
Organizational Affiliation: