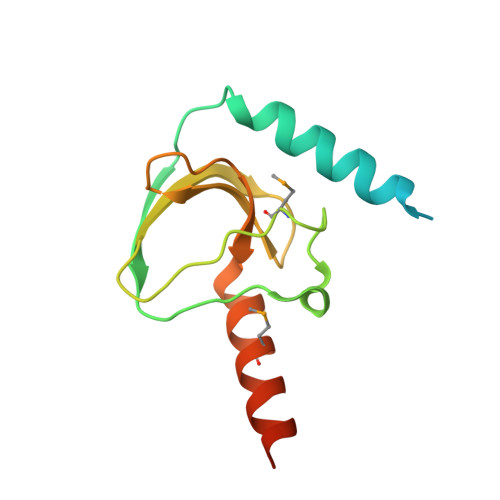
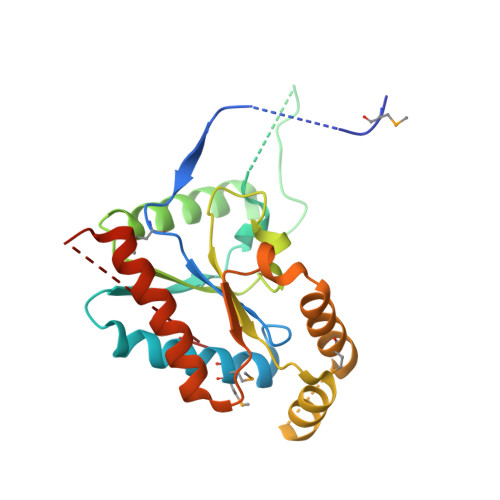
Structure of a complex between a voltage-gated calcium channel beta-subunit and an alpha-subunit domain.
Van Petegem, F., Clark, K.A., Chatelain, F.C., Minor, D.L.(2004) Nature 429: 671-675
- PubMed: 15141227
- DOI: https://doi.org/10.1038/nature02588
- Primary Citation of Related Structures:
1T0H, 1T0J - PubMed Abstract:
Voltage-gated calcium channels (Ca(V)s) govern muscle contraction, hormone and neurotransmitter release, neuronal migration, activation of calcium-dependent signalling cascades, and synaptic input integration. An essential Ca(V) intracellular protein, the beta-subunit (Ca(V)beta), binds a conserved domain (the alpha-interaction domain, AID) between transmembrane domains I and II of the pore-forming alpha(1) subunit and profoundly affects multiple channel properties such as voltage-dependent activation, inactivation rates, G-protein modulation, drug sensitivity and cell surface expression. Here, we report the high-resolution crystal structures of the Ca(V)beta2a conserved core, alone and in complex with the AID. Previous work suggested that a conserved region, the beta-interaction domain (BID), formed the AID-binding site; however, this region is largely buried in the Ca(V)beta core and is unavailable for protein-protein interactions. The structure of the AID-Ca(V)beta2a complex shows instead that Ca(V)beta2a engages the AID through an extensive, conserved hydrophobic cleft (named the alpha-binding pocket, ABP). The ABP-AID interaction positions one end of the Ca(V)beta near the intracellular end of a pore-lining segment, called IS6, that has a critical role in Ca(V) inactivation. Together, these data suggest that Ca(V)betas influence Ca(V) gating by direct modulation of IS6 movement within the channel pore.
- Cardiovascular Research Institute, Department of Biochemistry and Biophysics, University of California San Francisco, 513 Parnassus Avenue, Box 0130, San Francisco, California 94143, USA.
Organizational Affiliation: